Domain architectures of the Scm3p protein provide insights into centromere function and evolution
- PMID: 17704645
- PMCID: PMC2394858
- DOI: 10.4161/cc.6.20.4793
Domain architectures of the Scm3p protein provide insights into centromere function and evolution
Abstract
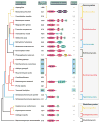
Recently, Scm3p has been shown to be a nonhistone component of centromeric chromatin that binds stoichiometrically to CenH3-H4 histones, and to be required for the assembly of kinetochores in Saccharomyces cerevisiae. Scm3p is conserved across fungi, and displays a remarkable variation in protein size, ranging from approximately 200 amino acids in S. cerevisiae to approximately 1300 amino acids in Neurospora crassa. This is primarily due a variable C-terminal segment that is linked to a conserved N-terminal, CenH3-interacting domain. We have discovered that the extended C-terminal region of Scm3p is strikingly characterized by lineage-specific fusions of single or multiple predicted DNA-binding domains different versions of the MYB and C2H2 zinc finger domains, AT-hooks, and a novel cysteine-rich metal-chelating cluster that are absent from the small versions of Scm3. Instead, S. cerevisiae point centromeres are recognized by components of the CBF3 DNA binding complex, which are conserved amongst close relatives of budding yeast, but are correspondingly absent from more distant fungi that possess regional centromeres. Hence, the C-terminal DNA binding motifs found in large Scm3p proteins may, along with CenH3, serve as a key epigenetic signal by recognizing and accommodating the lineage-specific diversity of centromere DNA in course of evolution.
Figures
References
-
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–102. - PubMed
-
- Henikoff S, Dalal Y. Centromeric chromatin: what makes it unique? Curr Opin Genet Dev. 2005;15:177–84. - PubMed
-
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
