Alkylation of prohibitin by cyclohexylphenyl-chloroethyl urea on an aspartyl residue is associated with cell cycle G(1) arrest in B16 cells
- PMID: 17704829
- PMCID: PMC2050821
- DOI: 10.1038/sj.bjp.0707415
Alkylation of prohibitin by cyclohexylphenyl-chloroethyl urea on an aspartyl residue is associated with cell cycle G(1) arrest in B16 cells
Abstract
Background and purpose: Phenyl-chloroethyl ureas (CEUs) are a class of anticancer drugs that mainly react with proteins. Two molecules of this family, cyclohexylphenyl-chloroethyl urea (CCEU) and iodophenyl-chloroethyl urea (ICEU) induced G(1)/S and G(2)/M cell cycle blocks, respectively. We hypothesised that these observations were linked to a differential protein alkylation pattern.
Experimental approach: Proteins from B16 cells incubated with [(14)C-urea]-CCEU and [(125)I]-ICEU were compared by 2D-analyses followed by MALDI-TOF identification of modified proteins and characterisation of the CCEU binding. Protein expression was investigated by Western blot analyses and cell cycle data were obtained by flow cytometry.
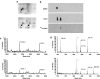
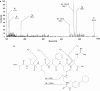
Key results: Several proteins (PDIA1, PDIA3, PDIA6, TRX, VDAC2) were alkylated by both ICEU and CCEU but beta-tubulin and prohibitin (PHB) were specifically alkylated by either ICEU or CCEU respectively. Specific alkylation of these two proteins might explain the observed difference in B16 cell cycle arrest in G(2) and G(1) phases respectively. Mass spectrometry studies on the alkylated prohibitin localised the modified peptide and identified Asp-40 as the target for CCEU. This alkylation induced an increased cellular content of PHB that should contribute to the accumulation of cells in G(1) phase.
Conclusions and implications: This study reinforces our findings that CEUs alkylate proteins through an ester linkage with an acidic amino acid and shows that PHB alkylation contributes to G(1)/S arrest in CCEU treated B16 cells. Modification of PHB status and/or activity is an open route for new cancer therapeutics.
Figures




Similar articles
-
Intramolecular cyclization of N-phenyl N'(2-chloroethyl)ureas leads to active N-phenyl-4,5-dihydrooxazol-2-amines alkylating β-tubulin Glu198 and prohibitin Asp40.Biochem Pharmacol. 2011 May 1;81(9):1116-23. doi: 10.1016/j.bcp.2011.02.014. Epub 2011 Mar 1. Biochem Pharmacol. 2011. PMID: 21371445
-
Selective alkylation of beta(II)-tubulin and thioredoxin-1 by structurally related subsets of aryl chloroethylureas leading to either anti-microtubules or redox modulating agents.Bioorg Med Chem. 2008 Aug 1;16(15):7277-90. doi: 10.1016/j.bmc.2008.06.028. Epub 2008 Jun 20. Bioorg Med Chem. 2008. PMID: 18617414
-
Alkylation potency and protein specificity of aromatic urea derivatives and bioisosteres as potential irreversible antagonists of the colchicine-binding site.Bioorg Med Chem. 2007 Jul 1;15(13):4456-69. doi: 10.1016/j.bmc.2007.04.028. Epub 2007 Apr 22. Bioorg Med Chem. 2007. PMID: 17498960
-
Prohibitin: a potential target for new therapeutics.Trends Mol Med. 2005 Apr;11(4):192-7. doi: 10.1016/j.molmed.2005.02.004. Trends Mol Med. 2005. PMID: 15823758 Review.
-
Urea derivatives as anticancer agents.Anticancer Agents Med Chem. 2009 May;9(4):471-80. doi: 10.2174/1871520610909040471. Anticancer Agents Med Chem. 2009. PMID: 19442045 Review.
Cited by
-
Synthesis, biological evaluation, and structure-activity relationships of novel substituted N-phenyl ureidobenzenesulfonate derivatives blocking cell cycle progression in S-phase and inducing DNA double-strand breaks.J Med Chem. 2012 Jul 12;55(13):6194-208. doi: 10.1021/jm3006492. Epub 2012 Jun 21. J Med Chem. 2012. PMID: 22694057 Free PMC article.
-
Design, synthesis, biological evaluation, and structure-activity relationships of substituted phenyl 4-(2-oxoimidazolidin-1-yl)benzenesulfonates as new tubulin inhibitors mimicking combretastatin A-4.J Med Chem. 2011 Jul 14;54(13):4559-80. doi: 10.1021/jm200488a. Epub 2011 Jun 13. J Med Chem. 2011. PMID: 21604746 Free PMC article.
References
-
- Bouchon B, Chambon C, Mounetou E, Papon J, Miot-Noirault E, Gaudreault RC, et al. Alkylation of beta-tubulin on Glu 198 by a microtubule disrupter. Mol Pharmacol. 2005;68:1415–1422. - PubMed
-
- Debiton E, Madelmont JC, Legault J, Barthomeuf C. Sanguinarine-induced apoptosis is associated with an early and severe cellular glutathione depletion. Cancer Chemother Pharmacol. 2003;51:474–482. - PubMed
-
- Fortin S, Moreau E, Patenaude A, Desjardins M, Lacroix J, Rousseau JL, et al. N-Phenyl-N′-(2-chloroethyl)ureas (CEU) as potential antineoplastic agents. Part 2: role of omega-hydroxyl group in the covalent binding to beta-tubulin. Bioorg Med Chem. 2007;15:1430–1438. - PubMed
-
- Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem. 2003;278:47853–47861. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

