Extensive parallelism in protein evolution
- PMID: 17705846
- PMCID: PMC2020468
- DOI: 10.1186/1745-6150-2-20
Extensive parallelism in protein evolution
Abstract
Background: Independently evolving lineages mostly accumulate different changes, which leads to their gradual divergence. However, parallel accumulation of identical changes is also common, especially in traits with only a small number of possible states.
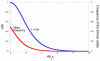
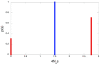
Results: We characterize parallelism in evolution of coding sequences in three four-species sets of genomes of mammals, Drosophila, and yeasts. Each such set contains two independent evolutionary paths, which we call paths I and II. An amino acid replacement which occurred along path I also occurs along path II with the probability 50-80% of that expected under selective neutrality. Thus, the per site rate of parallel evolution of proteins is several times higher than their average rate of evolution, but still lower than the rate of evolution of neutral sequences. This deficit may be caused by changes in the fitness landscape, leading to a replacement being possible along path I but not along path II. However, constant, weak selection assumed by the nearly neutral model of evolution appears to be a more likely explanation. Then, the average coefficient of selection associated with an amino acid replacement, in the units of the effective population size, must exceed approximately 0.4, and the fraction of effectively neutral replacements must be below approximately 30%. At a majority of evolvable amino acid sites, only a relatively small number of different amino acids is permitted.
Conclusion: High, but below-neutral, rates of parallel amino acid replacements suggest that a majority of amino acid replacements that occur in evolution are subject to weak, but non-trivial, selection, as predicted by Ohta's nearly-neutral theory.
Figures
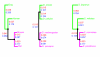
References
-
- Futuyma DJ. Evolution. Sunderland: Sinauer Associates; 2005.
-
- Li WH. Molecular Evolution. Sunderland: Sinauer Associates; 1997.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

