Optimization of candidate-gene SNP-genotyping by flexible oligonucleotide microarrays; analyzing variations in immune regulator genes of hay-fever samples
- PMID: 17705862
- PMCID: PMC1994959
- DOI: 10.1186/1471-2164-8-282
Optimization of candidate-gene SNP-genotyping by flexible oligonucleotide microarrays; analyzing variations in immune regulator genes of hay-fever samples
Abstract
Background: Genetic variants in immune regulator genes have been associated with numerous diseases, including allergies and cancer. Increasing evidence suggests a substantially elevated disease risk in individuals who carry a combination of disease-relevant single nucleotide polymorphisms (SNPs). For the genotyping of immune regulator genes, such as cytokines, chemokines and transcription factors, an oligonucleotide microarray for the analysis of 99 relevant SNPs was established. Since the microarray design was based on a platform that permits flexible in situ oligonucleotide synthesis, a set of optimally performing probes could be defined by a selection approach that combined computational and experimental aspects.
Results: While the in silico process eliminated 9% of the initial probe set, which had been picked purely on the basis of potential association with disease, the subsequent experimental validation excluded more than twice as many. The performance of the optimized microarray was demonstrated in a pilot study. The genotypes of 19 hay-fever patients (aged 40-44) with high IgE levels against inhalant antigens were compared to the results obtained with 19 age- and sex-matched controls. For several variants, allele-frequency differences of more than 10% were identified.
Conclusion: Based on the ability to improve empirically a chip design, the application of candidate-SNP typing represents a viable approach in the context of molecular epidemiological studies.
Figures

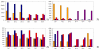


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

