Lessons learned from the initial sequencing of the pig genome: comparative analysis of an 8 Mb region of pig chromosome 17
- PMID: 17705864
- PMCID: PMC2374978
- DOI: 10.1186/gb-2007-8-8-r168
Lessons learned from the initial sequencing of the pig genome: comparative analysis of an 8 Mb region of pig chromosome 17
Abstract
Background: We describe here the sequencing, annotation and comparative analysis of an 8 Mb region of pig chromosome 17, which provides a useful test region to assess coverage and quality for the pig genome sequencing project. We report our findings comparing the annotation of draft sequence assembled at different depths of coverage.
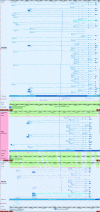
Results: Within this region we annotated 71 loci, of which 53 are orthologous to human known coding genes. When compared to the syntenic regions in human (20q13.13-q13.33) and mouse (chromosome 2, 167.5 Mb-178.3 Mb), this region was found to be highly conserved with respect to gene order. The most notable difference between the three species is the presence of a large expansion of zinc finger coding genes and pseudogenes on mouse chromosome 2 between Edn3 and Phactr3 that is absent from pig and human. All of our annotation has been made publicly available in the Vertebrate Genome Annotation browser, VEGA. We assessed the impact of coverage on sequence assembly across this region and found, as expected, that increased sequence depth resulted in fewer, longer contigs. One-third of our annotated loci could not be fully re-aligned back to the low coverage version of the sequence, principally because the transcripts are fragmented over several contigs.
Conclusion: We have demonstrated the considerable advantages of sequencing at increased read depths and discuss the implications that lower coverage sequence may have on subsequent comparative and functional studies, particularly those involving complex loci such as GNAS.
Figures


References
-
- Swine Genome Sequencing Consortium http://www.piggenome.org/index.php
-
- Wellcome Trust Sanger Institute Swine Genome Sequencing and Mapping http://www.sanger.ac.uk/Projects/S_scrofa/
-
- PreENSEMBL Pig Clone Map http://pre.ensembl.org/Sus_scrofa_map/index.html
-
- PreENSEMBL Pig Genome Sequence http://pre.ensembl.org/Sus_scrofa/index.html
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

