Molecular basis of tropomyosin binding to tropomodulin, an actin-capping protein
- PMID: 17706248
- PMCID: PMC2134803
- DOI: 10.1016/j.jmb.2007.05.084
Molecular basis of tropomyosin binding to tropomodulin, an actin-capping protein
Abstract
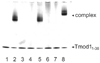
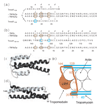
The tropomodulin (Tmod) family of proteins that cap the pointed, slow-growing end of actin filaments require tropomyosin (TM) for optimal function. Earlier studies identified two regions in Tmod1 that bind the N terminus of TM, though the ability of different isoforms to bind the two sites is controversial. We used model peptides to determine the affinity and define the specificity of the highly conserved N termini of three short, non-muscle TMs (alpha, gamma, delta-TM) for the two Tmod1 binding sites using circular dichroism spectroscopy, native gel electrophoresis, and chemical crosslinking. All TM peptides have high affinity for the second Tmod1 binding site (within residues 109-144; alpha-TM, 2.5 nM; gamma-TM, delta-TM, 40-90 nM), but differ >100-fold for the first site (residues 1-38; alpha-TM, 90 nM; undetectable at 10 microM, gamma-TM, delta-TM). Residue 14 (R in alpha; Q in gamma and delta) and, to a lesser extent, residue 4 (S in alpha; T in gamma and delta) are primarily responsible for the differences. The functional consequence of the sequence differences is reflected in more effective inhibition of actin filament elongation by full-length alpha-TMs than gamma-TM in the presence of Tmod1. The binding sites of the two Tmod1 peptides on a model TM peptide differ, as defined by comparing (15)N,(1)H HSQC spectra of a (15)N-labeled model TM peptide in both the absence and presence of Tmod1 peptide. The NMR and CD studies show that there is an increase in alpha-helix upon Tmod1-TM complex formation, indicating that intrinsically disordered regions of the two proteins become ordered upon binding. A model proposed for the binding of Tmod to actin and TM at the pointed end of the filament shows how the Tmod-TM accentuates the asymmetry of the pointed end and suggests how subtle differences among TM isoforms may modulate actin filament dynamics.
Figures




References
-
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. - PubMed
-
- Fowler VM. Regulation of actin filament length in erythrocytes and striated muscle. Curr Opin Cell Biol. 1996;8:86–96. - PubMed
-
- Littlefield R, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat Cell Biol. 2001;3:544–551. - PubMed
-
- Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

