Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus
- PMID: 17707229
- PMCID: PMC2034342
- DOI: 10.1016/j.molcel.2007.06.022
Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus
Abstract
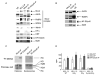
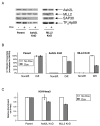
MLL-containing complexes methylate histone H3 at lysine 4 (H3K4) and have been implicated in the regulation of transcription. However, it is unclear how MLL complexes are targeted to specific gene loci. Here, we show that the MLL2 complex associates with the hematopoietic activator NF-E2 in erythroid cells and is important for H3K4 trimethylation and maximal levels of transcription at the beta-globin locus. Furthermore, recruitment of the MLL2 complex to the beta-globin locus is dependent upon NF-E2 and coincides spatio-temporally with NF-E2 binding during erythroid differentiation. Thus, a DNA-bound activator is important initially for guiding MLL2 to a particular genomic location. Interestingly, while the MLL2-associated subunit ASH2L is restricted to the beta-globin locus control region 38 kb upstream of the beta(maj)-globin gene, the MLL2 protein spreads across the beta-globin locus, suggesting a previously undefined mechanism by which an activator influences transcription and H3K4 trimethylation at a distance.
Figures


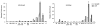

References
-
- Andrews NC. The NF-E2 transcription factor. Int J Biochem Cell Biol. 1998;30:429–432. - PubMed
-
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. 2005 - PubMed
-
- Genomic maps and comparative analysis of histone modifications in human and mouse. Cell . 120:169–181. - PubMed
-
- Brand M, Ranish JA, Kummer NT, Hamilton J, Igarashi K, Francastel C, Chi TH, Crabtree GR, Aebersold R, Groudine M. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol. 2004;11:73–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

