Modulation of p53 function by SET8-mediated methylation at lysine 382
- PMID: 17707234
- PMCID: PMC2693209
- DOI: 10.1016/j.molcel.2007.07.012
Modulation of p53 function by SET8-mediated methylation at lysine 382
Abstract
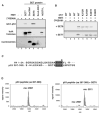
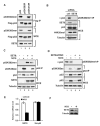
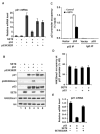
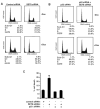
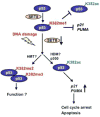
Reversible covalent methylation of lysine residues on histone proteins constitutes a principal molecular mechanism that links chromatin states to diverse biological outcomes. Recently, lysine methylation has been observed on nonhistone proteins, suggesting broad cellular roles for the enzymes generating and removing methyl moieties. Here we report that the lysine methyltransferase enzyme SET8/PR-Set7 regulates the tumor suppressor protein p53. We find that SET8 specifically monomethylates p53 at lysine 382 (p53K382me1). This methylation event robustly suppresses p53-mediated transcription activation of highly responsive target genes but has little influence on weak targets. Further, depletion of SET8 augments the proapoptotic and checkpoint activation functions of p53, and accordingly, SET8 expression is downregulated upon DNA damage. Together, our study identifies SET8 as a p53-modifying enzyme, identifies p53K382me1 as a regulatory posttranslational modification of p53, and begins to dissect how methylation may contribute to a dynamic posttranslational code that modulates distinct p53 functions.
Figures


References
-
- Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–2772. - PubMed
-
- Bannister AJ, Kouzarides T. Histone methylation: recognizing the methyl mark. Methods Enzymol. 2004;376:269–288. - PubMed
-
- Beisel C, Imhof A, Greene J, Kremmer E, Sauer F. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature. 2002;419:857–862. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

