The Drosophila ARC homolog regulates behavioral responses to starvation
- PMID: 17707655
- PMCID: PMC2094000
- DOI: 10.1016/j.mcn.2007.06.008
The Drosophila ARC homolog regulates behavioral responses to starvation
Abstract
The gene encoding dARC1, one of three Drosophila homologs of mammalian activity-regulated cytoskeleton-associated protein (ARC), is upregulated in both seizure and muscular hypercontraction mutants. In this study we generate a null mutant for dArc1 and show that this gene is not involved in synaptic plasticity at the larval neuromuscular junction or in formation or decay of short-term memory of courtship conditioning, but rather is a modifier of stress-induced behavior. dARC1 is expressed in a number of neurosecretory cells and mutants are starvation-resistant, exhibiting an increased time of survival in the absence of food. Starvation resistance is likely due to the fact that dArc1 mutants lack the normal hyperlocomotor response to starvation, which is almost universal in the animal kingdom. dARC1 acts in insulin-producing neurons of the pars intercerebralis to control this behavior, but does not appear to be a general regulator of insulin signaling. This suggests that there are multiple modes of communication between the pars and the ring gland that control starvation-induced behavioral responses.
Figures
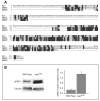
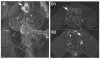
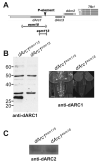


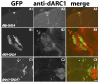


References
-
- Bader MF, Doussau F, Chasserot-Golaz S, Vitale N, Gasman S. Coupling actin and membrane dynamics during calcium-regulated exocytosis: a role for Rho and ARF GTPases. Biochim Biophys Acta. 2004;1742:37–49. - PubMed
-
- Berasi SP, Huard C, Li D, Shih HH, Sun Y, Zhong W, Paulsen JE, Brown EL, Gimeno RE, Martinez RV. Inhibition of gluconeogenesis through transcriptional activation of EGR1 and DUSP4 by AMP-activated kinase. J Biol Chem. 2006;281:27167–27177. - PubMed
-
- Beumont PJ, Arthur B, Russell JD, Touyz SW. Excessive physical activity in dieting disorder patients: proposals for a supervised exercise program. The International journal of eating disorders. 1994;15:21–36. - PubMed
-
- Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, Hafen E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865–875. - PubMed
-
- Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, Partridge L. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. 2005;102:3105–3110. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

