Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome
- PMID: 17708682
- PMCID: PMC1950163
- DOI: 10.1371/journal.pgen.0030136
Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome
Abstract
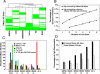
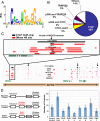
The identification of regulatory elements from different cell types is necessary for understanding the mechanisms controlling cell type-specific and housekeeping gene expression. Mapping DNaseI hypersensitive (HS) sites is an accurate method for identifying the location of functional regulatory elements. We used a high throughput method called DNase-chip to identify 3,904 DNaseI HS sites from six cell types across 1% of the human genome. A significant number (22%) of DNaseI HS sites from each cell type are ubiquitously present among all cell types studied. Surprisingly, nearly all of these ubiquitous DNaseI HS sites correspond to either promoters or insulator elements: 86% of them are located near annotated transcription start sites and 10% are bound by CTCF, a protein with known enhancer-blocking insulator activity. We also identified a large number of DNaseI HS sites that are cell type specific (only present in one cell type); these regions are enriched for enhancer elements and correlate with cell type-specific gene expression as well as cell type-specific histone modifications. Finally, we found that approximately 8% of the genome overlaps a DNaseI HS site in at least one the six cell lines studied, indicating that a significant percentage of the genome is potentially functional.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures




References
-
- Lemon B, Tjian R. Orchestrated response: A symphony of transcription factors for gene control. Genes Dev. 2000;14:2551–2569. - PubMed
-
- Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. - PubMed
-
- Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. - PubMed
-
- Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem Sci. 2000;25:99–104. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

