Quantitative genomics of locomotor behavior in Drosophila melanogaster
- PMID: 17708775
- PMCID: PMC2375002
- DOI: 10.1186/gb-2007-8-8-r172
Quantitative genomics of locomotor behavior in Drosophila melanogaster
Abstract
Background: Locomotion is an integral component of most animal behaviors, and many human health problems are associated with locomotor deficits. Locomotor behavior is a complex trait, with population variation attributable to many interacting loci with small effects that are sensitive to environmental conditions. However, the genetic basis of this complex behavior is largely uncharacterized.
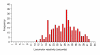
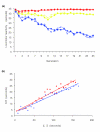
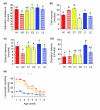
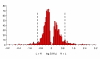
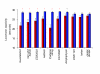
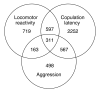
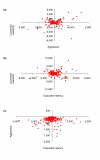
Results: We quantified locomotor behavior of Drosophila melanogaster in a large population of inbred lines derived from a single natural population, and derived replicated selection lines with different levels of locomotion. Estimates of broad-sense and narrow-sense heritabilities were 0.52 and 0.16, respectively, indicating substantial non-additive genetic variance for locomotor behavior. We used whole genome expression analysis to identify 1,790 probe sets with different expression levels between the selection lines when pooled across replicates, at a false discovery rate of 0.001. The transcriptional responses to selection for locomotor, aggressive and mating behavior from the same base population were highly overlapping, but the magnitude of the expression differences between selection lines for increased and decreased levels of behavior was uncorrelated. We assessed the locomotor behavior of ten mutations in candidate genes with altered transcript abundance between selection lines, and identified seven novel genes affecting this trait.
Conclusion: Expression profiling of genetically divergent lines is an effective strategy for identifying genes affecting complex behaviors, and reveals that a large number of pleiotropic genes exhibit correlated transcriptional responses to multiple behaviors.
Figures
References
-
- Chen Q, He Y, Yang K. Gene therapy for Parkinson's disease: progress and challenges. Curr Gene Therapy. 2005;5:71–80. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

