Spatial complexity and control of a bacterial cell cycle
- PMID: 17709236
- PMCID: PMC2716793
- DOI: 10.1016/j.copbio.2007.07.007
Spatial complexity and control of a bacterial cell cycle
Abstract
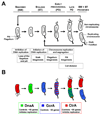
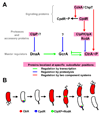
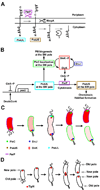
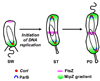
A major breakthrough in understanding the bacterial cell cycle is the discovery that bacteria exhibit a high degree of intracellular organization. Chromosomal loci and many protein complexes are positioned at particular subcellular sites. In this review, we examine recently discovered control mechanisms that make use of dynamically localized protein complexes to orchestrate the Caulobacter crescentus cell cycle. Protein localization, notably of signal transduction proteins, chromosome partition proteins, and proteases, serves to coordinate cell division with chromosome replication and cell differentiation. The developmental fate of daughter cells is decided before completion of cytokinesis, via the early establishment of cell polarity by the distribution of activated signaling proteins, bacterial cytoskeleton, and landmark proteins.
Figures
References
-
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. - PubMed
-
- Quon KC, Marczynski GT, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

