Advances in understanding the genetic basis of antimalarial drug resistance
- PMID: 17709280
- PMCID: PMC2080794
- DOI: 10.1016/j.mib.2007.07.007
Advances in understanding the genetic basis of antimalarial drug resistance
Abstract
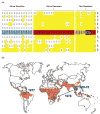
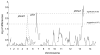
The acquisition of drug resistance by Plasmodium falciparum has severely curtailed global efforts to control malaria. Our ability to define resistance has been greatly enhanced by recent advances in Plasmodium genetics and genomics. Sequencing and microarray studies have identified thousands of polymorphisms in the P. falciparum genome, and linkage disequilibrium analyses have exploited these to rapidly identify known and novel loci that influence parasite susceptibility to antimalarials such as chloroquine, quinine, and sulfadoxine-pyrimethamine. Genetic approaches have also been designed to predict determinants of in vivo resistance to more recent first-line antimalarials such as the artemisinins. Transfection methodologies have defined the role of determinants including pfcrt, pfmdr1, and dhfr. This knowledge can be leveraged to develop more efficient methods of surveillance and treatment.
Figures


References
-
- Wellems TE, Plowe CV. Chloroquine-resistant malaria. J Infect Dis. 2001;184:770–776. - PubMed
-
- Jeffares DC, Pain A, Berry A, Cox AV, Stalker J, Ingle CE, Thomas A, Quail MA, Siebenthall K, Uhlemann AC, et al. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet. 2007;39:120–125. ●The authors describe the genome wide shotgun sequencing of a Ghanaian clinical isolate, the IT laboratory strain, and the related chimpanzee parasite P. reichenowi. They identify 27,000 nonredundant SNPs and a similar number of indels. The paper includes a comparison of protein evolutionary rates relative to their expression levels, developmental stage of expression and cellular localization. - PMC - PubMed
-
- Mu J, Awadalla P, Duan J, McGee KM, Keebler J, Seydel K, McVean GA, Su XZ. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet. 2007;39:126–130. ● These authors sequenced 3,539 genes (or about 65% of the total predicted genes in the P. falciparum genome) from the clones Dd2, Hb3, D10 and 7G8. They identify specific genes and genomic regions under diversifying selective pressure. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

