Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules
- PMID: 17709342
- PMCID: PMC2034483
- DOI: 10.1093/nar/gkm629
Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules
Abstract
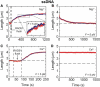
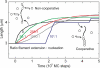
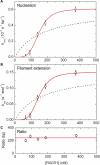
The human DNA repair protein RAD51 is the crucial component of helical nucleoprotein filaments that drive homologous recombination. The molecular mechanistic details of how this structure facilitates the requisite DNA strand rearrangements are not known but must involve dynamic interactions between RAD51 and DNA. Here, we report the real-time kinetics of human RAD51 filament assembly and disassembly on individual molecules of both single- and double-stranded DNA, as measured using magnetic tweezers. The relative rates of nucleation and filament extension are such that the observed filament formation consists of multiple nucleation events that are in competition with each other. For varying concentration of RAD51, a Hill coefficient of 4.3 +/- 0.5 is obtained for both nucleation and filament extension, indicating binding to dsDNA with a binding unit consisting of multiple (> or =4) RAD51 monomers. We report Monte Carlo simulations that fit the (dis)assembly data very well. The results show that, surprisingly, human RAD51 does not form long continuous filaments on DNA. Instead each nucleoprotein filament consists of a string of many small filament patches that are only a few tens of monomers long. The high flexibility and dynamic nature of this arrangement is likely to facilitate strand exchange.
Figures




References
-
- West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. - PubMed
-
- Wyman C, Kanaar R. Homologous recombination: Down to the wire. Curr. Biol. 2004;14:R629–R631. - PubMed
-
- Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 2004;11:791–796. - PubMed
-
- Yu X, VanLoock MS, Yang S, Reese JT, Egelman EH. What is the structure of the RecA-DNA filament? Curr. Protein Pept. Sc. 2004;5:73–79. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

