Transcriptome annotation using tandem SAGE tags
- PMID: 17709346
- PMCID: PMC2034470
- DOI: 10.1093/nar/gkm495
Transcriptome annotation using tandem SAGE tags
Abstract
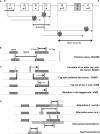
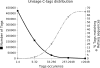
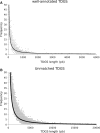
Analysis of several million expressed gene signatures (tags) revealed an increasing number of different sequences, largely exceeding that of annotated genes in mammalian genomes. Serial analysis of gene expression (SAGE) can reveal new Poly(A) RNAs transcribed from previously unrecognized chromosomal regions. However, conventional SAGE tags are too short to identify unambiguously unique sites in large genomes. Here, we design a novel strategy with tags anchored on two different restrictions sites of cDNAs. New transcripts are then tentatively defined by the two SAGE tags in tandem and by the spanning sequence read on the genome between these tagged sites. Having developed a new algorithm to locate these tag-delimited genomic sequences (TDGS), we first validated its capacity to recognize known genes and its ability to reveal new transcripts with two SAGE libraries built in parallel from a single RNA sample. Our algorithm proves fast enough to experiment this strategy at a large scale. We then collected and processed the complete sets of human SAGE tags to predict yet unknown transcripts. A cross-validation with tiling arrays data shows that 47% of these TDGS overlap transcriptional active regions. Our method provides a new and complementary approach for complex transcriptome annotation.
Figures


Similar articles
-
[Transcriptomes for serial analysis of gene expression].J Soc Biol. 2002;196(4):303-7. J Soc Biol. 2002. PMID: 12645300 Review. French.
-
Tag-based approaches for transcriptome research and genome annotation.Nat Methods. 2005 Jul;2(7):495-502. doi: 10.1038/nmeth768. Nat Methods. 2005. PMID: 15973418 Review.
-
Reverse serial analysis of gene expression (SAGE) characterization of orphan SAGE tags from human embryonic stem cells identifies the presence of novel transcripts and antisense transcription of key pluripotency genes.Stem Cells. 2006 May;24(5):1162-73. doi: 10.1634/stemcells.2005-0304. Epub 2006 Feb 2. Stem Cells. 2006. PMID: 16456128
-
Annotating nonspecific SAGE tags with microarray data.Genomics. 2006 Jan;87(1):173-80. doi: 10.1016/j.ygeno.2005.08.014. Epub 2005 Nov 28. Genomics. 2006. PMID: 16314072
-
Analysis of SAGE data in human platelets: features of the transcriptome in an anucleate cell.Thromb Haemost. 2006 Apr;95(4):643-51. Thromb Haemost. 2006. PMID: 16601835
Cited by
-
Combining DGE and RNA-sequencing data to identify new polyA+ non-coding transcripts in the human genome.Nucleic Acids Res. 2014 Mar;42(5):2820-32. doi: 10.1093/nar/gkt1300. Epub 2013 Dec 18. Nucleic Acids Res. 2014. PMID: 24357408 Free PMC article.
-
Using reads to annotate the genome: influence of length, background distribution, and sequence errors on prediction capacity.Nucleic Acids Res. 2009 Aug;37(15):e104. doi: 10.1093/nar/gkp492. Epub 2009 Jun 16. Nucleic Acids Res. 2009. PMID: 19531739 Free PMC article.
References
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. - PubMed
-
- Claverie JM. Fewer genes, more noncoding RNA. Science. 2005;309:1529–1530. - PubMed
-
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. - PubMed
-
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

