The birth of new exons: mechanisms and evolutionary consequences
- PMID: 17709368
- PMCID: PMC1986822
- DOI: 10.1261/rna.682507
The birth of new exons: mechanisms and evolutionary consequences
Abstract
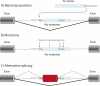
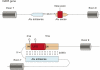
A significant amount of literature was dedicated to hypotheses concerning the origin of ancient introns and exons, but accumulating evidence indicates that new exons are also constantly being added to evolving genomes. Several mechanisms contribute to the creation of novel exons in metazoan genomes, including whole gene and single exon duplications, but perhaps the most intriguing are events of exonization, where intronic sequences become exons de novo. Exonizations of intronic sequences, particularly those originating from repetitive elements, are now widely documented in many genomes including human, mouse, dog, and fish. Such de novo appearance of exons is very frequently associated with alternative splicing, with the new exon-containing variant typically being the rare one. This allows the new variant to be evolutionarily tested without compromising the original one, and provides an evolutionary strategy for generation of novel functions with minimum damage to the existing functional repertoire. This review discusses the molecular mechanisms leading to exonization, its extent in vertebrate genomes, and its evolutionary implications.
Figures
Similar articles
-
Exonization of transposed elements: A challenge and opportunity for evolution.Biochimie. 2011 Nov;93(11):1928-34. doi: 10.1016/j.biochi.2011.07.014. Epub 2011 Jul 26. Biochimie. 2011. PMID: 21787833 Review.
-
Global analysis of exon creation versus loss and the role of alternative splicing in 17 vertebrate genomes.RNA. 2007 May;13(5):661-70. doi: 10.1261/rna.325107. Epub 2007 Mar 16. RNA. 2007. PMID: 17369312 Free PMC article.
-
Exon creation and establishment in human genes.Genome Biol. 2008;9(9):R141. doi: 10.1186/gb-2008-9-9-r141. Genome Biol. 2008. PMID: 18811936 Free PMC article.
-
Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss.Nat Genet. 2003 Jun;34(2):177-80. doi: 10.1038/ng1159. Nat Genet. 2003. PMID: 12730695
-
Modular assembly of genes and the evolution of new functions.Genetica. 2003 Jul;118(2-3):217-31. Genetica. 2003. PMID: 12868611 Review.
Cited by
-
Nothing in Evolution Makes Sense Except in the Light of Genomics: Read-Write Genome Evolution as an Active Biological Process.Biology (Basel). 2016 Jun 8;5(2):27. doi: 10.3390/biology5020027. Biology (Basel). 2016. PMID: 27338490 Free PMC article. Review.
-
Inosine in Biology and Disease.Genes (Basel). 2021 Apr 19;12(4):600. doi: 10.3390/genes12040600. Genes (Basel). 2021. PMID: 33921764 Free PMC article. Review.
-
Evolution of acetylcholinesterase and butyrylcholinesterase in the vertebrates: an atypical butyrylcholinesterase from the Medaka Oryzias latipes.PLoS One. 2011 Feb 25;6(2):e17396. doi: 10.1371/journal.pone.0017396. PLoS One. 2011. PMID: 21364766 Free PMC article.
-
Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities.Genome Biol. 2011 Jul 25;12(7):R68. doi: 10.1186/gb-2011-12-7-r68. Genome Biol. 2011. PMID: 21787409 Free PMC article.
-
Analysis of the canine brain transcriptome with an emphasis on the hypothalamus and cerebral cortex.Mamm Genome. 2013 Dec;24(11-12):484-99. doi: 10.1007/s00335-013-9480-0. Epub 2013 Nov 8. Mamm Genome. 2013. PMID: 24202129
References
-
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. - PubMed
-
- Ast, G. How did alternative splicing evolve? Nat. Rev. Genet. 2004;5:773–782. - PubMed
-
- Barton, R.M., Worman, H.J. Prenylated prelamin A interacts with Narf, a novel nuclear protein. J. Biol. Chem. 1999;274:30008–30018. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
