Two domains of cytotoxic necrotizing factor type 1 bind the cellular receptor, laminin receptor precursor protein
- PMID: 17709415
- PMCID: PMC2168285
- DOI: 10.1128/IAI.00075-07
Two domains of cytotoxic necrotizing factor type 1 bind the cellular receptor, laminin receptor precursor protein
Abstract
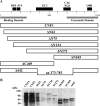
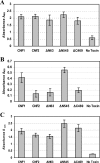
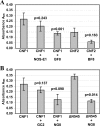
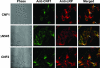
Cytotoxic necrotizing factor type 1 (CNF1) and CNF2 are highly homologous toxins that are produced by certain pathogenic strains of Escherichia coli. These 1,014-amino-acid toxins catalyze the deamidation of a specific glutamine residue in RhoA, Rac1, and Cdc42 and consist of a putative N-terminal binding domain, a transmembrane region, and a C-terminal catalytic domain. To define the regions of CNF1 that are responsible for binding of the toxin to its cellular receptor, the laminin receptor precursor protein (LRP), a series of CNF1 truncated toxins were characterized and assessed for toxin binding. In particular, three truncated toxins, DeltaN63, DeltaN545, and DeltaC469, retained conformational integrity and in vitro enzymatic activity and were immunologically reactive against a panel of anti-CNF1 monoclonal antibodies (MAbs). Based on a comparison of these truncated toxins with wild-type CNF1 and CNF2 in LRP and HEp-2 cell binding assays and in MAb and LRP competitive binding inhibition assays and based on the results of confocal microscopy, we concluded that CNF1 contains two major binding regions: one located within the N terminus, which contained amino acids 135 to 164, and one which resided in the C terminus and included amino acids 683 to 730. The data further indicate that CNF1 can bind to an additional receptor(s) on HEp-2 cells and that LRP can also serve as a cellular receptor for CNF2.
Figures


References
-
- Aktories, K. 1997. Rho proteins: targets for bacterial toxins. Trends Microbiol. 5:282-288. - PubMed
-
- Boquet, P. 2001. The cytotoxic necrotizing factor 1 (CNF1) from Escherichia coli. Toxicon 39:1673-1680. - PubMed
-
- Buetow, L., G. Flatau, K. Chiu, P. Boquet, and P. Ghosh. 2001. Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1. Nat. Struct. Biol. 8:584-588. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

