OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L
- PMID: 17709429
- PMCID: PMC2064540
- DOI: 10.1083/jcb.200704110
OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L
Abstract
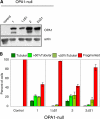
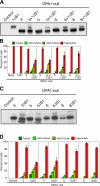
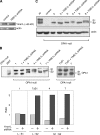
OPA1, a dynamin-related guanosine triphosphatase mutated in dominant optic atrophy, is required for the fusion of mitochondria. Proteolytic cleavage by the mitochondrial processing peptidase generates long isoforms from eight messenger RNA (mRNA) splice forms, whereas further cleavages at protease sites S1 and S2 generate short forms. Using OPA1-null cells, we developed a cellular system to study how individual OPA1 splice forms function in mitochondrial fusion. Only mRNA splice forms that generate a long isoform in addition to one or more short isoforms support substantial mitochondrial fusion activity. On their own, long and short OPA1 isoforms have little activity, but, when coexpressed, they functionally complement each other. Loss of mitochondrial membrane potential destabilizes the long isoforms and enhances the cleavage of OPA1 at S1 but not S2. Cleavage at S2 is regulated by the i-AAA protease Yme1L. Our results suggest that mammalian cells have multiple pathways to control mitochondrial fusion through regulation of the spectrum of OPA1 isoforms.
Figures


References
-
- Alexander, C., M. Votruba, U.E. Pesch, D.L. Thiselton, S. Mayer, A. Moore, M. Rodriguez, U. Kellner, B. Leo-Kottler, G. Auburger, et al. 2000. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26:211–215. - PubMed
-
- Arnold, I., and T. Langer. 2002. Membrane protein degradation by AAA proteases in mitochondria. Biochim. Biophys. Acta. 1592:89–96. - PubMed
-
- Chan, D.C. 2006. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 22:79–99. - PubMed
-
- Chen, H., A. Chomyn, and D.C. Chan. 2005. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 280:26185–26192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

