Regulation of the gonadal transcriptome during sex determination and testis morphogenesis: comparative candidate genes
- PMID: 17709564
- PMCID: PMC8260008
- DOI: 10.1530/REP-06-0341
Regulation of the gonadal transcriptome during sex determination and testis morphogenesis: comparative candidate genes
Abstract
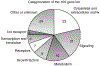
Gene expression profiles during sex determination and gonadal differentiation were investigated to identify new potential regulatory factors. Embryonic day 13 (E13), E14, and E16 rat testes and ovaries were used for microarray analysis, as well as E13 testis organ cultures that undergo testis morphogenesis and develop seminiferous cords in vitro. A list of 109 genes resulted from a selective analysis for genes present in male gonadal development and with a 1.5-fold change in expression between E13 and E16. Characterization of these 109 genes potentially important for testis development revealed that cytoskeletal-associated proteins, extracellular matrix factors, and signaling factors were highly represented. Throughout the developmental period (E13-E16), sex-enriched transcripts were more prevalent in the male with 34 of the 109 genes having testis-enriched expression during sex determination. In ovaries, the total number of transcripts with a 1.5-fold change in expression between E13 and E16 was similar to the testis, but none of those genes were both ovary enriched and regulated during the developmental period. Genes conserved in sex determination were identified by comparing changing transcripts in the rat analysis herein, to transcripts altered in previously published mouse studies of gonadal sex determination. A comparison of changing mouse and rat transcripts identified 43 genes with species conservation in sex determination and testis development. Profiles of gene expression during E13-E16 rat testis and ovary development are presented and candidate genes for involvement in sex determination and testis differentiation are identified. Analysis of cellular pathways did not reveal any specific pathways involving multiple candidate genes. However, the genes and gene network identified influence numerous cellular processes with cellular differentiation, proliferation, focal contact, RNA localization, and development being predominant.
Figures


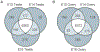



Similar articles
-
Role of neurotropins in rat embryonic testis morphogenesis (cord formation).Biol Reprod. 2000 Jan;62(1):132-42. doi: 10.1095/biolreprod62.1.132. Biol Reprod. 2000. PMID: 10611077
-
Inhibition of platelet-derived growth factor actions in the embryonic testis influences normal cord development and morphology.Biol Reprod. 2002 Mar;66(3):745-53. doi: 10.1095/biolreprod66.3.745. Biol Reprod. 2002. PMID: 11870082
-
Analysis of Functional cis-Regulatory Elements Reveals Novel Transcriptional Regulatory Mechanisms in Gonadal Development.Sex Dev. 2024;18(1-6):14-26. doi: 10.1159/000543594. Epub 2025 Jan 20. Sex Dev. 2024. PMID: 39832493
-
Genetic programs that regulate testicular and ovarian development.Mol Cell Endocrinol. 2007 Feb;265-266:3-9. doi: 10.1016/j.mce.2006.12.029. Epub 2007 Jan 8. Mol Cell Endocrinol. 2007. PMID: 17208359 Review.
-
Mammalian sex determination and gonad development.Curr Top Dev Biol. 2013;106:89-121. doi: 10.1016/B978-0-12-416021-7.00003-1. Curr Top Dev Biol. 2013. PMID: 24290348 Review.
Cited by
-
SRY directly regulates the neurotrophin 3 promoter during male sex determination and testis development in rats.Biol Reprod. 2011 Aug;85(2):277-84. doi: 10.1095/biolreprod.110.090282. Epub 2011 Apr 20. Biol Reprod. 2011. PMID: 21508350 Free PMC article.
-
Expression of miRNAs in ovine fetal gonads: potential role in gonadal differentiation.Reprod Biol Endocrinol. 2011 Jan 11;9:2. doi: 10.1186/1477-7827-9-2. Reprod Biol Endocrinol. 2011. PMID: 21223560 Free PMC article.
-
Basic helix-loop-helix transcription factor TCF21 is a downstream target of the male sex determining gene SRY.PLoS One. 2011;6(5):e19935. doi: 10.1371/journal.pone.0019935. Epub 2011 May 17. PLoS One. 2011. PMID: 21637323 Free PMC article.
-
Retinoic Acid Induced Protein 14 (Rai14) is dispensable for mouse spermatogenesis.PeerJ. 2021 Feb 19;9:e10847. doi: 10.7717/peerj.10847. eCollection 2021. PeerJ. 2021. PMID: 33643708 Free PMC article.
-
Characterization of gonadal transcriptomes from Nile tilapia (Oreochromis niloticus) reveals differentially expressed genes.PLoS One. 2013 May 3;8(5):e63604. doi: 10.1371/journal.pone.0063604. Print 2013. PLoS One. 2013. PMID: 23658843 Free PMC article.
References
-
- Anand-Srivastava MB 2005. Natriuretic peptide receptor-C signaling and regulation. Peptides 26 1044–1059. - PubMed
-
- Asirvatham AJ, Schmidt M, Gao B & Chaudhary J 2006. Androgens regulate the immune/inflammatory response and cell survival pathways in rat ventral prostate epithelial cells. Endocrinology 147 257–271. - PubMed
-
- Bartoloni L, Wattenhofer M, Kudoh J, Berry A, Shibuya K, Kawasaki K, Wang J, Asakawa S, Talior T, Bonne-Tamir B, Rossier C, Michaud J, McCabe ERB, Minoshima S, Shimizu N, Scott HS & Antonarakis SE 2000. Cloning and characterization of a putative human glycerol 3-phosphate permease gene (SLC37A1 or G3PP) on 21q22.3: mutation analysis in two candidate phenotypes, DFNB10 and a glycerol kinase deficiency. Genomics 70 190–200. - PubMed
-
- Berta P, Hawkins JR, Sinclair AH, Taylor A, Griffiths BL, Goodfellow PN & Fellous M 1990. Genetic evidence equating SRYand the testis-determining factor. Nature 348 448–450. - PubMed
-
- Beverdam A & Koopman P 2006. Expression profiling of purified mouse gonadal somatic cells during the critical time window of sex determination reveals novel candidate genes for human sexual dysgenesis syndromes. Human Molecular Genetics 15 417–431. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

