Mendel's green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway
- PMID: 17709752
- PMCID: PMC1955798
- DOI: 10.1073/pnas.0705521104
Mendel's green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway
Abstract
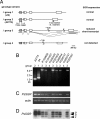
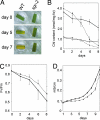
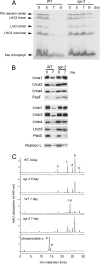
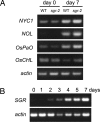
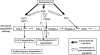
Mutants that retain greenness of leaves during senescence are known as "stay-green" mutants. The most famous stay-green mutant is Mendel's green cotyledon pea, one of the mutants used in determining the law of genetics. Pea plants homozygous for this recessive mutation (known as i at present) retain greenness of the cotyledon during seed maturation and of leaves during senescence. We found tight linkage between the I locus and stay-green gene originally found in rice, SGR. Molecular analysis of three i alleles including one with no SGR expression confirmed that the I gene encodes SGR in pea. Functional analysis of sgr mutants in pea and rice further revealed that leaf functionality is lowered despite a high chlorophyll a (Chl a) and chlorophyll b (Chl b) content in the late stage of senescence, suggesting that SGR is primarily involved in Chl degradation. Consistent with this observation, a wide range of Chl-protein complexes, but not the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) large subunit, were shown to be more stable in sgr than wild-type plants. The expression of OsCHL and NYC1, which encode the first enzymes in the degrading pathways of Chl a and Chl b, respectively, was not affected by sgr in rice. The results suggest that SGR might be involved in activation of the Chl-degrading pathway during leaf senescence through translational or posttranslational regulation of Chl-degrading enzymes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

