C. elegans agrin is expressed in pharynx, IL1 neurons and distal tip cells and does not genetically interact with genes involved in synaptogenesis or muscle function
- PMID: 17710131
- PMCID: PMC1939731
- DOI: 10.1371/journal.pone.0000731
C. elegans agrin is expressed in pharynx, IL1 neurons and distal tip cells and does not genetically interact with genes involved in synaptogenesis or muscle function
Abstract
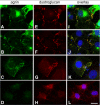
Agrin is a basement membrane protein crucial for development and maintenance of the neuromuscular junction in vertebrates. The C. elegans genome harbors a putative agrin gene agr-1. We have cloned the corresponding cDNA to determine the primary structure of the protein and expressed its recombinant fragments to raise specific antibodies. The domain organization of AGR-1 is very similar to the vertebrate orthologues. C. elegans agrin contains a signal sequence for secretion, seven follistatin domains, three EGF-like repeats and two laminin G domains. AGR-1 loss of function mutants did not exhibit any overt phenotypes and did not acquire resistance to the acetylcholine receptor agonist levamisole. Furthermore, crossing them with various mutants for components of the dystrophin-glycoprotein complex with impaired muscle function did not lead to an aggravation of the phenotypes. Promoter-GFP translational fusion as well as immunostaining of worms revealed expression of agrin in buccal epithelium and the protein deposition in the basal lamina of the pharynx. Furthermore, dorsal and ventral IL1 head neurons and distal tip cells of the gonad arms are sources of agrin production, but no expression was detectable in body muscles or in the motoneurons innervating them. Recombinant worm AGR-1 fragment is able to cluster vertebrate dystroglycan in cultured cells, implying a conservation of this interaction, but since neither of these proteins is expressed in muscle of C. elegans, this interaction may be required in different tissues. The connections between muscle cells and the basement membrane, as well as neuromuscular junctions, are structurally distinct between vertebrates and nematodes.
Conflict of interest statement
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

