Classification of a moderately oxygen-tolerant isolate from baby faeces as Bifidobacterium thermophilum
- PMID: 17711586
- PMCID: PMC2045100
- DOI: 10.1186/1471-2180-7-79
Classification of a moderately oxygen-tolerant isolate from baby faeces as Bifidobacterium thermophilum
Abstract
Background: Bifidobacteria are found at varying prevalence in human microbiota and seem to play an important role in the human gastrointestinal tract (GIT). Bifidobacteria are highly adapted to the human GIT which is reflected in the genome sequence of a Bifidobacterim longum isolate. The competitiveness against other bacteria is not fully understood yet but may be related to the production of antimicrobial compounds such as bacteriocins. In a previous study, 34 Bifidobacterium isolates have been isolated from baby faeces among which six showed proteinaceous antilisterial activity against Listeria monocytogenes. In this study, one of these isolates, RBL67, was further identified and characterized.
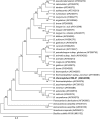
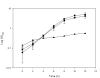
Results: Bifidobacterium isolate RBL67 was classified and characterized using a polyphasic approach. RBL67 was classified as Bifidobacterium thermophilum based on phenotypic and DNA-DNA hybridization characteristics, although 16S rDNA analyses and partial groEL sequences showed higher homology with B. thermacidophilum subsp. porcinum and B. thermacidophilum subsp. thermacidophilum, respectively. RBL67 was moderately oxygen-tolerant and was able to grow at pH 4 and at a temperature of 47 degrees C.
Conclusion: In order to assign RBL67 to a species, a polyphasic approach was used. This resulted in the classification of RBL67 as a Bifidobacterium thermophilum strain. To our knowledge, this is the first report about B. thermophilum isolated from baby faeces since the B. thermophilum strains were related to ruminants and swine faeces before. B. thermophilum was previously only isolated from animal sources and was therefore suggested to be used as differential species between animal and human contamination. Our findings may disapprove this suggestion and further studies are now conducted to determine whether B. thermophilum is distributed broader in human faeces. Furthermore, the postulated differentiation between human and animal strains by growth above 45 degrees C is no longer valid since B. thermophilum is able to grow at 47 degrees C. In our study, 16S rDNA and partial groEL sequence analysis were not able to clearly assign RBL67 to a species and were contradictory. Our study suggests that partial groEL sequences may not be reliable as a single tool for species differentiation.
Figures


References
-
- Tissier MH. La réaction chromophile d'Escherich et Bacterium Coli. C R Seances Soc Biol Fil. 1899;51:943–945.
-
- Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen M-C, Desiere F, Bork P, Delley M, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci USA. 2002;99:14422–14427. doi: 10.1073/pnas.212527599. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

