HIV-1 protease and reverse transcriptase control the architecture of their nucleocapsid partner
- PMID: 17712401
- PMCID: PMC1940317
- DOI: 10.1371/journal.pone.0000669
HIV-1 protease and reverse transcriptase control the architecture of their nucleocapsid partner
Erratum in
- PLoS One. 2014;9(8):e105187
Abstract
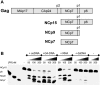
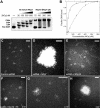
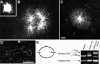
The HIV-1 nucleocapsid is formed during protease (PR)-directed viral maturation, and is transformed into pre-integration complexes following reverse transcription in the cytoplasm of the infected cell. Here, we report a detailed transmission electron microscopy analysis of the impact of HIV-1 PR and reverse transcriptase (RT) on nucleocapsid plasticity, using in vitro reconstitutions. After binding to nucleic acids, NCp15, a proteolytic intermediate of nucleocapsid protein (NC), was processed at its C-terminus by PR, yielding premature NC (NCp9) followed by mature NC (NCp7), through the consecutive removal of p6 and p1. This allowed NC co-aggregation with its single-stranded nucleic-acid substrate. Examination of these co-aggregates for the ability of RT to catalyse reverse transcription showed an effective synthesis of double-stranded DNA that, remarkably, escaped from the aggregates more efficiently with NCp7 than with NCp9. These data offer a compelling explanation for results from previous virological studies that focused on i) Gag processing leading to nucleocapsid condensation, and ii) the disappearance of NCp7 from the HIV-1 pre-integration complexes. We propose that HIV-1 PR and RT, by controlling the nucleocapsid architecture during the steps of condensation and dismantling, engage in a successive nucleoprotein-remodelling process that spatiotemporally coordinates the pre-integration steps of HIV-1. Finally we suggest that nucleoprotein remodelling mechanisms are common features developed by mobile genetic elements to ensure successful replication.
Conflict of interest statement
Figures
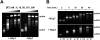

Similar articles
-
A protein ballet around the viral genome orchestrated by HIV-1 reverse transcriptase leads to an architectural switch: from nucleocapsid-condensed RNA to Vpr-bridged DNA.Virus Res. 2013 Feb;171(2):287-303. doi: 10.1016/j.virusres.2012.09.008. Epub 2012 Sep 24. Virus Res. 2013. PMID: 23017337 Free PMC article.
-
Formation of immature and mature genomic RNA dimers in wild-type and protease-inactive HIV-1: differential roles of the Gag polyprotein, nucleocapsid proteins NCp15, NCp9, NCp7, and the dimerization initiation site.Virology. 2010 Nov 25;407(2):225-36. doi: 10.1016/j.virol.2010.08.013. Epub 2010 Sep 9. Virology. 2010. PMID: 20828778
-
Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein.J Mol Biol. 2006 Dec 1;364(3):496-511. doi: 10.1016/j.jmb.2006.08.065. Epub 2006 Aug 30. J Mol Biol. 2006. PMID: 17020765
-
Overview of the Nucleic-Acid Binding Properties of the HIV-1 Nucleocapsid Protein in Its Different Maturation States.Viruses. 2020 Sep 29;12(10):1109. doi: 10.3390/v12101109. Viruses. 2020. PMID: 33003650 Free PMC article. Review.
-
The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance.Curr HIV Res. 2004 Jan;2(1):79-92. doi: 10.2174/1570162043485022. Curr HIV Res. 2004. PMID: 15053342 Review.
Cited by
-
Nucleocapsid Protein: A Desirable Target for Future Therapies Against HIV-1.Curr Top Microbiol Immunol. 2015;389:53-92. doi: 10.1007/82_2015_433. Curr Top Microbiol Immunol. 2015. PMID: 25749978 Free PMC article. Review.
-
Reaction-diffusion basis of retroviral infectivity.Philos Trans A Math Phys Eng Sci. 2016 Nov 13;374(2080):20160148. doi: 10.1098/rsta.2016.0148. Philos Trans A Math Phys Eng Sci. 2016. PMID: 27698042 Free PMC article.
-
Blocking premature reverse transcription fails to rescue the HIV-1 nucleocapsid-mutant replication defect.Retrovirology. 2011 Jun 17;8:46. doi: 10.1186/1742-4690-8-46. Retrovirology. 2011. PMID: 21682883 Free PMC article.
-
A protein ballet around the viral genome orchestrated by HIV-1 reverse transcriptase leads to an architectural switch: from nucleocapsid-condensed RNA to Vpr-bridged DNA.Virus Res. 2013 Feb;171(2):287-303. doi: 10.1016/j.virusres.2012.09.008. Epub 2012 Sep 24. Virus Res. 2013. PMID: 23017337 Free PMC article.
-
Investigating the cellular distribution and interactions of HIV-1 nucleocapsid protein by quantitative fluorescence microscopy.PLoS One. 2015 Feb 27;10(2):e0116921. doi: 10.1371/journal.pone.0116921. eCollection 2015. PLoS One. 2015. PMID: 25723396 Free PMC article.
References
-
- Pettit SC, Sheng N, Tritch R, Erickson-Viitanen S, Swanstrom R. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv Exp Med Biol. 1998;436:15–25. - PubMed
-
- Arhel N, Genovesio A, Kim KA, Miko S, Perret E, et al. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nat Methods. 2006;3:817–824. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

