Sequential logic model deciphers dynamic transcriptional control of gene expressions
- PMID: 17712424
- PMCID: PMC1945082
- DOI: 10.1371/journal.pone.0000776
Sequential logic model deciphers dynamic transcriptional control of gene expressions
Abstract
Background: Cellular signaling involves a sequence of events from ligand binding to membrane receptors through transcription factors activation and the induction of mRNA expression. The transcriptional-regulatory system plays a pivotal role in the control of gene expression. A novel computational approach to the study of gene regulation circuits is presented here.
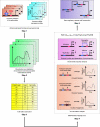
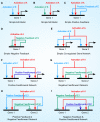
Methodology: Based on the concept of finite state machine, which provides a discrete view of gene regulation, a novel sequential logic model (SLM) is developed to decipher control mechanisms of dynamic transcriptional regulation of gene expressions. The SLM technique is also used to systematically analyze the dynamic function of transcriptional inputs, the dependency and cooperativity, such as synergy effect, among the binding sites with respect to when, how much and how fast the gene of interest is expressed.
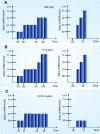
Principal findings: SLM is verified by a set of well studied expression data on endo16 of Strongylocentrotus purpuratus (sea urchin) during the embryonic midgut development. A dynamic regulatory mechanism for endo16 expression controlled by three binding sites, UI, R and Otx is identified and demonstrated to be consistent with experimental findings. Furthermore, we show that during transition from specification to differentiation in wild type endo16 expression profile, SLM reveals three binary activities are not sufficient to explain the transcriptional regulation of endo16 expression and additional activities of binding sites are required. Further analyses suggest detailed mechanism of R switch activity where indirect dependency occurs in between UI activity and R switch during specification to differentiation stage.
Conclusions/significance: The sequential logic formalism allows for a simplification of regulation network dynamics going from a continuous to a discrete representation of gene activation in time. In effect our SLM is non-parametric and model-independent, yet providing rich biological insight. The demonstration of the efficacy of this approach in endo16 is a promising step for further application of the proposed method.
Conflict of interest statement
Figures






Similar articles
-
Brn1/2/4, the predicted midgut regulator of the endo16 gene of the sea urchin embryo.Dev Biol. 2005 May 15;281(2):286-98. doi: 10.1016/j.ydbio.2005.02.034. Dev Biol. 2005. PMID: 15893979
-
Cis-regulatory logic in the endo16 gene: switching from a specification to a differentiation mode of control.Development. 2001 Mar;128(5):617-29. doi: 10.1242/dev.128.5.617. Development. 2001. PMID: 11171388
-
Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation.Development. 2003 Sep;130(17):4187-99. doi: 10.1242/dev.00611. Development. 2003. PMID: 12874137
-
Deciphering the underlying mechanism of specification and differentiation: the sea urchin gene regulatory network.Sci STKE. 2006 Nov 14;2006(361):pe47. doi: 10.1126/stke.3612006pe47. Sci STKE. 2006. PMID: 17106076 Review.
-
Modeling DNA sequence-based cis-regulatory gene networks.Dev Biol. 2002 Jun 1;246(1):2-13. doi: 10.1006/dbio.2002.0617. Dev Biol. 2002. PMID: 12027430 Review.
Cited by
-
Systems Biology Strategy Reveals PKCδ is Key for Sensitizing TRAIL-Resistant Human Fibrosarcoma.Front Immunol. 2015 Jan 5;5:659. doi: 10.3389/fimmu.2014.00659. eCollection 2014. Front Immunol. 2015. PMID: 25601862 Free PMC article.
-
Robustness under functional constraint: the genetic network for temporal expression in Drosophila neurogenesis.PLoS Comput Biol. 2010 Apr 29;6(4):e1000760. doi: 10.1371/journal.pcbi.1000760. PLoS Comput Biol. 2010. PMID: 20454677 Free PMC article.
-
Signaling flux redistribution at toll-like receptor pathway junctions.PLoS One. 2008;3(10):e3430. doi: 10.1371/journal.pone.0003430. Epub 2008 Oct 17. PLoS One. 2008. PMID: 18927610 Free PMC article.
-
Multiscale models of angiogenesis.IEEE Eng Med Biol Mag. 2009 Mar-Apr;28(2):14-31. doi: 10.1109/MEMB.2009.931791. IEEE Eng Med Biol Mag. 2009. PMID: 19349248 Free PMC article. Review.
References
-
- Wolf DM, Eeckman FH. On the relationship between genomic regulatory element organization and gene regulatory dynamics. J Theor Biol. 1998;195:167–186. - PubMed
-
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. - PubMed
-
- Hasty J, McMillen D, Isaacs F, Collins JJ. Computational studies of gene regulatory networks: in numero molecular biology. Nat Rev Genet. 2001;2:268–279. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

