Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii
- PMID: 17714582
- PMCID: PMC2089061
- DOI: 10.1186/1471-2229-7-45
Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii
Abstract
Background: The holoparasitic plant genus Cuscuta comprises species with photosynthetic capacity and functional chloroplasts as well as achlorophyllous and intermediate forms with restricted photosynthetic activity and degenerated chloroplasts. Previous data indicated significant differences with respect to the plastid genome coding capacity in different Cuscuta species that could correlate with their photosynthetic activity. In order to shed light on the molecular changes accompanying the parasitic lifestyle, we sequenced the plastid chromosomes of the two species Cuscuta reflexa and Cuscuta gronovii. Both species are capable of performing photosynthesis, albeit with varying efficiencies. Together with the plastid genome of Epifagus virginiana, an achlorophyllous parasitic plant whose plastid genome has been sequenced, these species represent a series of progression towards total dependency on the host plant, ranging from reduced levels of photosynthesis in C. reflexa to a restricted photosynthetic activity and degenerated chloroplasts in C. gronovii to an achlorophyllous state in E. virginiana.
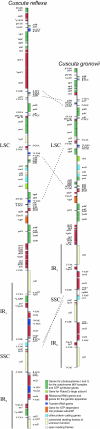
Results: The newly sequenced plastid genomes of C. reflexa and C. gronovii reveal that the chromosome structures are generally very similar to that of non-parasitic plants, although a number of species-specific insertions, deletions (indels) and sequence inversions were identified. However, we observed a gradual adaptation of the plastid genome to the different degrees of parasitism. The changes are particularly evident in C. gronovii and include (a) the parallel losses of genes for the subunits of the plastid-encoded RNA polymerase and the corresponding promoters from the plastid genome, (b) the first documented loss of the gene for a putative splicing factor, MatK, from the plastid genome and (c) a significant reduction of RNA editing.
Conclusion: Overall, the comparative genomic analysis of plastid DNA from parasitic plants indicates a bias towards a simplification of the plastid gene expression machinery as a consequence of an increasing dependency on the host plant. A tentative assignment of the successive events in the adaptation of the plastid genomes to parasitism can be inferred from the current data set. This includes (1) a loss of non-coding regions in photosynthetic Cuscuta species that has resulted in a condensation of the plastid genome, (2) the simplification of plastid gene expression in species with largely impaired photosynthetic capacity and (3) the deletion of a significant part of the genetic information, including the information for the photosynthetic apparatus, in non-photosynthetic parasitic plants.
Figures


References
-
- Panda MM, Choudhury NK. Effect of irradiance and nutrients on chlorophyll and carotenoid content and Hill reaction activity in Cuscuta reflexa. Photosynthetica. 1992;26:585–592.
-
- Sherman TD, Pettigraw WT, Vaughn K. Structural and immunological characterization of the Cuscuta pentagona L. chloroplast. Plant Cell Physiol. 1999;40:592–603.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

