Histone modifications in Trypanosoma brucei
- PMID: 17714803
- PMCID: PMC2012948
- DOI: 10.1016/j.molbiopara.2007.07.005
Histone modifications in Trypanosoma brucei
Abstract
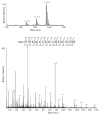
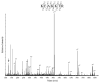
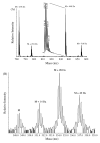
Several biological processes in Trypanosoma brucei are affected by chromatin structure, including gene expression, cell cycle regulation, and life-cycle stage differentiation. In Saccharomyces cerevisiae and other organisms, chromatin structure is dependent upon posttranslational modifications of histones, which have been mapped in detail. The tails of the four core histones of T. brucei are highly diverged from those of mammals and yeasts, so sites of potential modification cannot be reliably inferred, and no cross-species antibodies are available to map the modifications. We therefore undertook an extensive survey to identify posttranslational modifications by Edman degradation and mass spectrometry. Edman analysis showed that the N-terminal alanine of H2A, H2B, and H4 could be monomethylated. We found that the histone H4 N-terminus is heavily modified, while, in contrast to other organisms, the histone H2A and H2B N-termini have relatively few modifications. Histone H3 appears to have a number of modifications at the N-terminus, but we were unable to assign many of these to a specific amino acid. Therefore, we focused our efforts on uncovering modification states of H4. We discuss the potential relevance of these modifications.
Figures



References
-
- Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–83. - PubMed
-
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–52. - PubMed
-
- Brower-Toland B, Wacker DA, Fulbright RM, Lis JT, Kraus WL, Wang MD. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J Mol Biol. 2005;346:135–46. - PubMed
-
- Kuo MH, vom Baur E, Struhl K, Allis CD. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol Cell. 2000;6:1309–20. - PubMed
-
- Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4:276–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

