Crystal structure of the incretin-bound extracellular domain of a G protein-coupled receptor
- PMID: 17715056
- PMCID: PMC1955799
- DOI: 10.1073/pnas.0706404104
Crystal structure of the incretin-bound extracellular domain of a G protein-coupled receptor
Abstract
Incretins, endogenous polypeptide hormones released in response to food intake, potentiate insulin secretion from pancreatic beta cells after oral glucose ingestion (the incretin effect). This response is signaled by the two peptide hormones glucose-dependent insulinotropic polypeptide (GIP) (also known as gastric inhibitory polypeptide) and glucagon-like peptide 1 through binding and activation of their cognate class 2 G protein-coupled receptors (GPCRs). Because the incretin effect is lost or significantly reduced in patients with type 2 diabetes mellitus, glucagon-like peptide 1 and GIP have attracted considerable attention for their potential in antidiabetic therapy. A paucity of structural information precludes a detailed understanding of the processes of hormone binding and receptor activation, hampering efforts to develop novel pharmaceuticals. Here we report the crystal structure of the complex of human GIP receptor extracellular domain (ECD) with its agonist, the incretin GIP(1-42). The hormone binds in an alpha-helical conformation in a surface groove of the ECD largely through hydrophobic interactions. The N-terminal ligand residues would remain free to interact with other parts of the receptor. Thermodynamic data suggest that binding is concomitant with structural organization of the hormone, resulting in a complex mode of receptor-ligand recognition. The presentation of a well structured, alpha-helical ligand by the ECD is expected to be conserved among other hormone receptors of this class.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


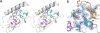

References
-
- Fehmann HC, Göke R, Göke B. Endocr Rev. 1995;16:390–410. - PubMed
-
- Drucker DJ. Cell Metab. 2006;3:153–165. - PubMed
-
- Nauck MA, Baller B, Meier JJ. Diabetes. 2004;53(Suppl 3):S190–S196. - PubMed
-
- O'Harte FP, Gray AM, Flatt PR. J Endocrinol. 1998;156:237–243. - PubMed
-
- Xie D, Cheng H, Hamrick M, Zhong Q, Ding KH, Correa D, Williams S, Mulloy A, Bollag W, Bollag RJ, et al. Bone. 2005;37:759–769. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

