Novel high-resolution characterization of ancient DNA reveals C > U-type base modification events as the sole cause of post mortem miscoding lesions
- PMID: 17715147
- PMCID: PMC2034480
- DOI: 10.1093/nar/gkm588
Novel high-resolution characterization of ancient DNA reveals C > U-type base modification events as the sole cause of post mortem miscoding lesions
Abstract
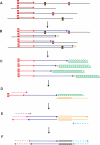
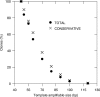
Ancient DNA (aDNA) research has long depended on the power of PCR to amplify trace amounts of surviving genetic material from preserved specimens. While PCR permits specific loci to be targeted and amplified, in many ways it can be intrinsically unsuited to damaged and degraded aDNA templates. PCR amplification of aDNA can produce highly-skewed distributions with significant contributions from miscoding lesion damage and non-authentic sequence artefacts. As traditional PCR-based approaches have been unable to fully resolve the molecular nature of aDNA damage over many years, we have developed a novel single primer extension (SPEX)-based approach to generate more accurate sequence information. SPEX targets selected template strands at defined loci and can generate a quantifiable redundancy of coverage; providing new insights into the molecular nature of aDNA damage and fragmentation. SPEX sequence data reveals inherent limitations in both traditional and metagenomic PCR-based approaches to aDNA, which can make current damage analyses and correct genotyping of ancient specimens problematic. In contrast to previous aDNA studies, SPEX provides strong quantitative evidence that C > U-type base modifications are the sole cause of authentic endogenous damage-derived miscoding lesions. This new approach could allow ancient specimens to be genotyped with unprecedented accuracy.
Figures


References
-
- Haak W, Forster P, Bramanti B, Matsumura S, Brandt G, Tanzer M, Villems R, Renfrew C, Gronenborn D, et al. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science. 2005;310:1016–1018. - PubMed
-
- Ritchie PA, Millar CD, Gibb GC, Baroni C, Lambert DM. Ancient DNA enables timing of the pleistocene origin and holocene expansion of two adelie penguin lineages in antarctica. Mol. Biol. Evol. 2004;21:240–248. - PubMed
-
- Shapiro B, Drummond AJ, Rambaut A, Wilson MC, Matheus PE, Sher AV, Pybus OG, Gilbert MT, Barnes I, et al. Rise and fall of the Beringian steppe bison. Science. 2004;306:1561–1565. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

