Polyomaviruses of birds: etiologic agents of inflammatory diseases in a tumor virus family
- PMID: 17715213
- PMCID: PMC2168798
- DOI: 10.1128/JVI.01178-07
Polyomaviruses of birds: etiologic agents of inflammatory diseases in a tumor virus family
Figures
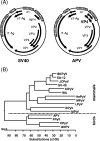
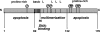
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

