Novel role for EKLF in megakaryocyte lineage commitment
- PMID: 17715392
- PMCID: PMC2190608
- DOI: 10.1182/blood-2007-03-082065
Novel role for EKLF in megakaryocyte lineage commitment
Abstract
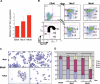
Megakaryocytes and erythroid cells are thought to derive from a common progenitor during hematopoietic differentiation. Although a number of transcriptional regulators are important for this process, they do not explain the bipotential result. We now show by gain- and loss-of-function studies that erythroid Krüppel-like factor (EKLF), a transcription factor whose role in erythroid gene regulation is well established, plays an unexpected directive role in the megakaryocyte lineage. EKLF inhibits the formation of megakaryocytes while at the same time stimulating erythroid differentiation. Quantitative examination of expression during hematopoiesis shows that, unlike genes whose presence is required for establishment of both lineages, EKLF is uniquely down-regulated in megakaryocytes after formation of the megakaryocyte-erythroid progenitor. Expression profiling and molecular analyses support these observations and suggest that megakaryocytic inhibition is achieved, at least in part, by EKLF repression of Fli-1 message levels.
Figures


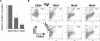



References
-
- Orkin SH. Diversification of haematopoietic stem cells to specific lineages. Nat Rev Genet. 2000;1:57–64. - PubMed
-
- Weissman IL, Anderson DJ, Gage F. Stem and progenitor cells: origins, phenotypes, lineage commitments, and transdifferentiations. Annu Rev Cell Dev Biol. 2001;17:387–403. - PubMed
-
- Kondo M, Wagers AJ, Manz MG, et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. - PubMed
-
- Traver D, Miyamoto T, Christensen J, Iwasaki-Arai J, Akashi K, Weissman IL. Fetal liver myelopoiesis occurs through distinct, prospectively isolatable progenitor subsets. Blood. 2001;98:627–635. - PubMed
-
- Adolfsson J, Mansson R, Buza-Vidas N, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

