Diversity in penaeidin antimicrobial peptide form and function
- PMID: 17716729
- PMCID: PMC2245800
- DOI: 10.1016/j.dci.2007.06.009
Diversity in penaeidin antimicrobial peptide form and function
Abstract
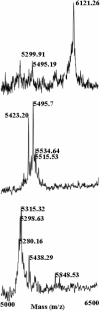
Penaeidins are a diverse family of two-domain antimicrobial peptides expressed in shrimp. Variation in penaeidin sequence results in functional diversity, which was discovered using synthetic reproductions of native penaeidins. An isoform of penaeidin class 3 from Litopenaeus setiferus (Litset Pen3-4) was synthesized using native ligation and compared directly with the synthetic penaeidin class 4 known to be expressed in the same organism. New antimicrobial activity data are included in this review that emphasize differences in effectiveness that are apparent from a direct comparison of two classes. A novel approach to intact penaeidin analysis is presented in the form of Fourier Transform Ion-Cyclotron Resonance Mass Spectrometry, which has implications for the identification of individual penaeidin isoforms without chemical modification or enzymatic cleavage. The new information included in this review helps gather the perspective on relevance of penaeidin diversity to antimicrobial function, the use of synthetic peptides as tools to evaluate specific immune functions and the application of high mass resolution, top-down sequencing methods to the intact analysis of individual penaeidin isoforms.
Figures






References
-
- Braff MH, Gallo RL. Antimicrobial peptides: an essential component of the skin defensive barrier. Curr Top Microbiol Immunol. 2006;306:91–110. - PubMed
-
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–95. - PubMed
-
- Bulet P, Hetru C, Dimarcq JL, Hoffmann D. Antimicrobial peptides in insects; structure and function. Dev Comp Immunol. 1999;23:329–44. - PubMed
-
- Dimarcq JL, Bulet P, Hetru C, Hoffmann J. Cysteine-rich antimicrobial peptides in invertebrates. Biopolymers. 1998;47:465–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

