Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence
- PMID: 17717001
- PMCID: PMC2034460
- DOI: 10.1093/nar/gkm579
Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence
Abstract
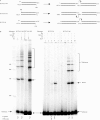
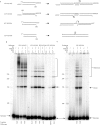
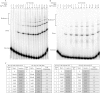
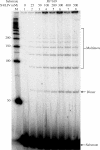
The double-strand DNA break repair pathway, non-homologous DNA end joining (NHEJ), is distinctive for the flexibility of its nuclease, polymerase and ligase activities. Here we find that the joining of ends by XRCC4-ligase IV is markedly influenced by the terminal sequence, and a steric hindrance model can account for this. XLF (Cernunnos) stimulates the joining of both incompatible DNA ends and compatible DNA ends at physiologic concentrations of Mg2+, but only of incompatible DNA ends at higher concentrations of Mg2+, suggesting charge neutralization between the two DNA ends within the ligase complex. XRCC4-DNA ligase IV has the distinctive ability to ligate poly-dT single-stranded DNA and long dT overhangs in a Ku- and XLF-independent manner, but not other homopolymeric DNA. The dT preference of the ligase is interesting given the sequence bias of the NHEJ polymerase. These distinctive properties of the XRCC4-DNA ligase IV complex explain important aspects of its in vivo roles.
Figures
References
-
- Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair. 2006;5:1234–1245. - PubMed
-
- Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv. Immunol. 2005;86:43–112. - PubMed
-
- Schwarz K, Ma Y, Pannicke U, Lieber MR. Human severe combined immune deficiency. Bioessays. 2003;25:1061–1070. - PubMed
-
- Moshous D, Callebaut I, Chasseval RD, Corneo B, Cavazzana-Calvo M, Diest FL, Tezcan I, Sanal O, Bertrand Y, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. - PubMed
-
- Callebaut I, Malivert L, Fischer A, Mornon JP, Revy P, Villartay JPD. Cernunnos interacts with the XRCC4/DNA-ligase IV complex and is homologous to the yeast nonhomologous end-joining factor NEJ1. J. Biol. Chem. 2006;281:13857–13860. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

