A novel olfactory receptor gene family in teleost fish
- PMID: 17717047
- PMCID: PMC1987349
- DOI: 10.1101/gr.6553207
A novel olfactory receptor gene family in teleost fish
Abstract
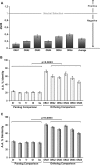
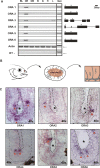
While for two of three mammalian olfactory receptor families (OR and V2R) ortholog teleost families have been identified, the third family (V1R) has been thought to be represented by a single, closely linked gene pair. We identified four further V1R-like genes in every teleost species analyzed (Danio rerio, Gasterosteus aculeatus, Oryzias latipes, Tetraodon nigroviridis, Takifugu rubripes). In the phylogenetic analysis these ora genes (olfactory receptor class A-related) form a single clade, which includes the entire mammalian V1R superfamily. Homologies are much lower in paralogs than in orthologs, indicating that all six family members are evolutionarily much older than the speciation events in the teleost lineage analyzed here. These ora genes are under strong negative selection, as evidenced by very small d(N)/d(S) values in comparisons between orthologs. A pairwise configuration in the phylogenetic tree suggests the existence of three ancestral Ora subclades, one of which has been lost in amphibia, and a further one in mammals. Unexpectedly, two ora genes exhibit a highly conserved multi-exonic structure and four ora genes are organized in closely linked gene pairs across all fish species studied. All ora genes are expressed specifically in the olfactory epithelium of zebrafish, in sparse cells within the sensory surface, consistent with the expectation for olfactory receptors. The ora gene repertoire is highly conserved across teleosts, in striking contrast to the frequent species-specific expansions observed in tetrapod, especially mammalian V1Rs, possibly reflecting a major shift in gene regulation as well as gene function upon the transition to tetrapods.
Figures




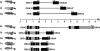
Similar articles
-
The molecular evolution of teleost olfactory receptor gene families.Results Probl Cell Differ. 2009;47:37-55. doi: 10.1007/400_2008_11. Results Probl Cell Differ. 2009. PMID: 18956167 Review.
-
Conservation of a vitellogenin gene cluster in oviparous vertebrates and identification of its traces in the platypus genome.Gene. 2008 Apr 30;413(1-2):76-82. doi: 10.1016/j.gene.2008.02.001. Epub 2008 Feb 9. Gene. 2008. PMID: 18343608
-
Olfactory expression of a single and highly variable V1r pheromone receptor-like gene in fish species.Proc Natl Acad Sci U S A. 2005 Apr 12;102(15):5489-94. doi: 10.1073/pnas.0402581102. Epub 2005 Apr 4. Proc Natl Acad Sci U S A. 2005. PMID: 15809442 Free PMC article.
-
Divergent evolution among teleost V1r receptor genes.PLoS One. 2007 Apr 18;2(4):e379. doi: 10.1371/journal.pone.0000379. PLoS One. 2007. PMID: 17440615 Free PMC article.
-
Identification of olfactory receptor genes in Atlantic salmon Salmo salar.J Fish Biol. 2012 Jul;81(2):559-75. doi: 10.1111/j.1095-8649.2012.03368.x. J Fish Biol. 2012. PMID: 22803724 Review.
Cited by
-
Loss of olfaction in sea snakes provides new perspectives on the aquatic adaptation of amniotes.Proc Biol Sci. 2019 Sep 11;286(1910):20191828. doi: 10.1098/rspb.2019.1828. Epub 2019 Sep 11. Proc Biol Sci. 2019. PMID: 31506057 Free PMC article.
-
The evolutionary origins of the vertebrate olfactory system.Open Biol. 2020 Dec;10(12):200330. doi: 10.1098/rsob.200330. Epub 2020 Dec 23. Open Biol. 2020. PMID: 33352063 Free PMC article. Review.
-
Positive Darwinian selection in the singularly large taste receptor gene family of an 'ancient' fish, Latimeria chalumnae.BMC Genomics. 2014 Aug 5;15(1):650. doi: 10.1186/1471-2164-15-650. BMC Genomics. 2014. PMID: 25091523 Free PMC article.
-
Coelacanth genomes reveal signatures for evolutionary transition from water to land.Genome Res. 2013 Oct;23(10):1740-8. doi: 10.1101/gr.158105.113. Epub 2013 Jul 22. Genome Res. 2013. PMID: 23878157 Free PMC article.
-
Family structure and phylogenetic analysis of odorant receptor genes in the large yellow croaker (Larimichthys crocea).BMC Evol Biol. 2011 Aug 11;11:237. doi: 10.1186/1471-2148-11-237. BMC Evol Biol. 2011. PMID: 21834959 Free PMC article.
References
-
- Boschat C., Pelofi C., Randin O., Roppolo D., Luscher C., Broillet M.C., Rodriguez I., Pelofi C., Randin O., Roppolo D., Luscher C., Broillet M.C., Rodriguez I., Randin O., Roppolo D., Luscher C., Broillet M.C., Rodriguez I., Roppolo D., Luscher C., Broillet M.C., Rodriguez I., Luscher C., Broillet M.C., Rodriguez I., Broillet M.C., Rodriguez I., Rodriguez I. Pheromone detection mediated by a V1r vomeronasal receptor. Nat. Neurosci. 2002;5:1261–1262. - PubMed
-
- Bryson-Richardson R.J., Logan D.W., Currie P.D., Jackson I.J., Logan D.W., Currie P.D., Jackson I.J., Currie P.D., Jackson I.J., Jackson I.J. Large-scale analysis of gene structure in rhodopsin-like GPCRs: Evidence for widespread loss of an ancient intron. Gene. 2004;338:15–23. - PubMed
-
- Buck L., Axel R., Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65:175–187. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
