Genetic analysis of the histidine utilization (hut) genes in Pseudomonas fluorescens SBW25
- PMID: 17717196
- PMCID: PMC1950622
- DOI: 10.1534/genetics.107.075713
Genetic analysis of the histidine utilization (hut) genes in Pseudomonas fluorescens SBW25
Abstract
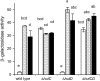
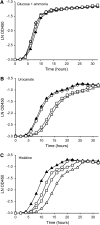
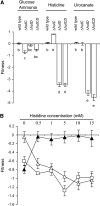
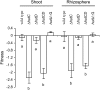
The histidine utilization (hut) locus of Pseudomonas fluorescens SBW25 confers the ability to utilize histidine as a sole carbon and nitrogen source. Genetic analysis using a combination of site-directed mutagenesis and chromosomally integrated lacZ fusions showed the hut locus to be composed of 13 genes organized in 3 transcriptional units: hutF, hutCD, and 10 genes from hutU to hutG (which includes 2 copies of hutH, 1 of which is nonfunctional). Inactivation of hutF eliminated the ability to grow on histidine, indicating that SBW25 degrades histidine by the five-step enzymatic pathway. The 3 hut operons are negatively regulated by the HutC repressor with urocanate (the first intermediate of the histidine degradation pathway) as the physiological inducer. 5'-RACE analysis of transcriptional start sites revealed involvement of both sigma(54) (for the hutU-G operon) and sigma(70) (for hutF); the involvement of sigma(54) was experimentally demonstrated. CbrB (an enhancer binding protein for sigma(54) recruitment) was required for bacterial growth on histidine, indicating positive control of hut gene expression by CbrB. Recognition that a gene (named hutD) encoding a widely distributed conserved hypothetical protein is transcribed along with hutC led to analysis of its role. Mutational and gene fusion studies showed that HutD functions independently of HutC. Growth and fitness assays in laboratory media and on sugar beet seedlings suggest that HutD acts as a governor that sets an upper bound to the level of hut activity.
Figures

Similar articles
-
Regulation of the histidine utilization (hut) system in bacteria.Microbiol Mol Biol Rev. 2012 Sep;76(3):565-84. doi: 10.1128/MMBR.00014-12. Microbiol Mol Biol Rev. 2012. PMID: 22933560 Free PMC article. Review.
-
Global Regulatory Roles of the Histidine-Responsive Transcriptional Repressor HutC in Pseudomonas fluorescens SBW25.J Bacteriol. 2020 Jun 9;202(13):e00792-19. doi: 10.1128/JB.00792-19. Print 2020 Jun 9. J Bacteriol. 2020. PMID: 32291279 Free PMC article.
-
Organization and multiple regulation of histidine utilization genes in Pseudomonas putida.J Bacteriol. 1988 Sep;170(9):4272-9. doi: 10.1128/jb.170.9.4272-4279.1988. J Bacteriol. 1988. PMID: 2842309 Free PMC article.
-
Dual involvement of CbrAB and NtrBC in the regulation of histidine utilization in Pseudomonas fluorescens SBW25.Genetics. 2008 Jan;178(1):185-95. doi: 10.1534/genetics.107.081984. Genetics. 2008. PMID: 18202367 Free PMC article.
-
The histidine utilization (hut) genes of Pseudomonas fluorescens SBW25 are active on plant surfaces, but are not required for competitive colonization of sugar beet seedlings.Microbiology (Reading). 2006 Jun;152(Pt 6):1867-1875. doi: 10.1099/mic.0.28731-0. Microbiology (Reading). 2006. PMID: 16735749
Cited by
-
Regulation of the histidine utilization (hut) system in bacteria.Microbiol Mol Biol Rev. 2012 Sep;76(3):565-84. doi: 10.1128/MMBR.00014-12. Microbiol Mol Biol Rev. 2012. PMID: 22933560 Free PMC article. Review.
-
Natural and engineered hydroxyectoine production based on the Pseudomonas stutzeri ectABCD-ask gene cluster.Appl Environ Microbiol. 2011 Feb;77(4):1368-74. doi: 10.1128/AEM.02124-10. Epub 2010 Dec 17. Appl Environ Microbiol. 2011. PMID: 21169432 Free PMC article.
-
Pseudomonas aeruginosa supports the survival of Prevotella melaninogenica in a cystic fibrosis lung polymicrobial community through metabolic cross-feeding.bioRxiv [Preprint]. 2024 Oct 21:2024.10.21.619475. doi: 10.1101/2024.10.21.619475. bioRxiv. 2024. PMID: 39484496 Free PMC article. Preprint.
-
AmyR is a novel negative regulator of amylovoran production in Erwinia amylovora.PLoS One. 2012;7(9):e45038. doi: 10.1371/journal.pone.0045038. Epub 2012 Sep 18. PLoS One. 2012. PMID: 23028751 Free PMC article.
-
Genotypic and phenotypic analyses reveal distinct population structures and ecotypes for sugar beet-associated Pseudomonas in Oxford and Auckland.Ecol Evol. 2020 May 11;10(12):5963-5975. doi: 10.1002/ece3.6334. eCollection 2020 Jun. Ecol Evol. 2020. PMID: 32607204 Free PMC article.
References
-
- Atkinson, M. R., N. Pattaramanon and A. J. Ninfa, 2002. Governor of the glnAp2 promoter of Escherichia coli. Mol. Microbiol. 46: 1247–1257. - PubMed
-
- Chasin, L. A., and B. Magasanik, 1968. Induction and repression of the histidine-degrading enzymes of Bacillus subtilis. J. Biol. Chem. 243: 5165–5178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

