Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma
- PMID: 17717605
- PMCID: PMC1950459
- DOI: 10.1172/JCI31457
Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma
Abstract
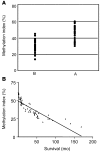
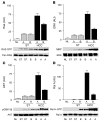
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide, accounting for an estimated 600,000 deaths annually. Aberrant methylation, consisting of DNA hypomethylation and/or promoter gene CpG hypermethylation, is implicated in the development of a variety of solid tumors, including HCC. We analyzed the global levels of DNA methylation as well as the methylation status of 105 putative tumor suppressor genes and found that the extent of genome-wide hypomethylation and CpG hypermethylation correlates with biological features and clinical outcome of HCC patients. We identified activation of Ras and downstream Ras effectors (ERK, AKT, and RAL) due to epigenetic silencing of inhibitors of the Ras pathway in all HCC. Further, selective inactivation of SPRY1 and -2, DAB2, and SOCS4 and -5 genes and inhibitors of angiogenesis (BNIP3, BNIP3L, IGFBP3, and EGLN2) was associated with poor prognosis. Importantly, several epigenetically silenced putative tumor suppressor genes found in HCC were also inactivated in the nontumorous liver. Our results assign both therapeutic and chemopreventive significance to methylation patterns in human HCC and open the possibility of using molecular targets, including those identified in this study, to effectively inhibit HCC development and progression.
Figures




References
-
- Bruix J., Boix L., Sala M., Llovet J.M. Focus on hepatocellular carcinoma. Cancer Cell. 2004;5:215–219. - PubMed
-
- Thorgeirsson S.S., Grisham J.W. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 2002;31:339–346. - PubMed
-
- Baylin S.B. Mechanisms underlying epigenetically mediated gene silencing in cancer. Semin. Cancer Biol. 2002;12:331–337. - PubMed
-
- Wilson A.S., Power B.E., Molloy P.L. DNA hypomethylation and human diseases. Biochim. Biophys. Acta. 2007;1775:138–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

