An AICD-based functional screen to identify APP metabolism regulators
- PMID: 17718916
- PMCID: PMC2071909
- DOI: 10.1186/1750-1326-2-15
An AICD-based functional screen to identify APP metabolism regulators
Abstract
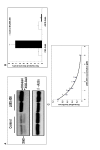
Background: A central event in Alzheimer's disease (AD) is the regulated intramembraneous proteolysis of the beta-amyloid precursor protein (APP), to generate the beta-amyloid (Abeta) peptide and the APP intracellular domain (AICD). Abeta is the major component of amyloid plaques and AICD displays transcriptional activation properties. We have taken advantage of AICD transactivation properties to develop a genetic screen to identify regulators of APP metabolism. This screen relies on an APP-Gal4 fusion protein, which upon normal proteolysis, produces AICD-Gal4. Production of AICD-Gal4 induces Gal4-UAS driven luciferase expression. Therefore, when regulators of APP metabolism are modulated, luciferase expression is altered.
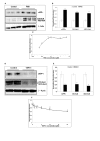
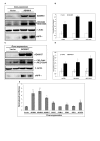
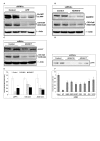
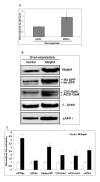
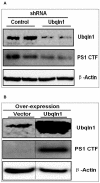
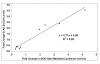
Results: To validate this experimental approach we modulated alpha-, beta-, and gamma-secretase levels and activities. Changes in AICD-Gal4 levels as measured by Western blot analysis were strongly and significantly correlated to the observed changes in AICD-Gal4 mediated luciferase activity. To determine if a known regulator of APP trafficking/maturation and Presenilin1 endoproteolysis could be detected using the AICD-Gal4 mediated luciferase assay, we knocked-down Ubiquilin 1 and observed decreased luciferase activity. We confirmed that Ubiquilin 1 modulated AICD-Gal4 levels by Western blot analysis and also observed that Ubiquilin 1 modulated total APP levels, the ratio of mature to immature APP, as well as PS1 endoproteolysis.
Conclusion: Taken together, we have shown that this screen can identify known APP metabolism regulators that control proteolysis, intracellular trafficking, maturation and levels of APP and its proteolytic products. We demonstrate for the first time that Ubiquilin 1 regulates APP metabolism in the human neuroblastoma cell line, SH-SY5Y.
Figures



Similar articles
-
Ubiquilin-1 modulates γ-secretase-mediated ε-site cleavage in neuronal cells.Biochemistry. 2013 Jun 4;52(22):3899-912. doi: 10.1021/bi400138p. Epub 2013 May 21. Biochemistry. 2013. PMID: 23663107
-
Purification and aggregation of the amyloid precursor protein intracellular domain.J Vis Exp. 2012 Aug 28;(66):e4204. doi: 10.3791/4204. J Vis Exp. 2012. PMID: 22952038 Free PMC article.
-
APP intracellular domain derived from amyloidogenic β- and γ-secretase cleavage regulates neprilysin expression.Front Aging Neurosci. 2015 May 19;7:77. doi: 10.3389/fnagi.2015.00077. eCollection 2015. Front Aging Neurosci. 2015. PMID: 26074811 Free PMC article.
-
The amyloid precursor protein intracellular domain (AICD) as modulator of gene expression, apoptosis, and cytoskeletal dynamics-relevance for Alzheimer's disease.Prog Neurobiol. 2008 Aug;85(4):393-406. doi: 10.1016/j.pneurobio.2008.05.002. Epub 2008 Jul 7. Prog Neurobiol. 2008. PMID: 18603345 Review.
-
beta-Amyloid precursor protein metabolism: focus on the functions and degradation of its intracellular domain.Pharmacol Res. 2010 Oct;62(4):308-17. doi: 10.1016/j.phrs.2010.05.002. Epub 2010 May 16. Pharmacol Res. 2010. PMID: 20561999 Review.
Cited by
-
Degradation and inhibition of epigenetic regulatory protein BRD4 exacerbate Alzheimer's disease-related neuropathology in cell models.J Biol Chem. 2022 Apr;298(4):101794. doi: 10.1016/j.jbc.2022.101794. Epub 2022 Mar 3. J Biol Chem. 2022. PMID: 35248531 Free PMC article.
-
Drosophila lilliputian is required for proneural gene expression in retinal development.Dev Dyn. 2012 Mar;241(3):553-62. doi: 10.1002/dvdy.23738. Epub 2012 Jan 25. Dev Dyn. 2012. PMID: 22275119 Free PMC article.
-
Synaptic NMDA receptor activation stimulates alpha-secretase amyloid precursor protein processing and inhibits amyloid-beta production.J Neurosci. 2009 Apr 8;29(14):4442-60. doi: 10.1523/JNEUROSCI.6017-08.2009. J Neurosci. 2009. PMID: 19357271 Free PMC article.
-
Loss of function of ATXN1 increases amyloid beta-protein levels by potentiating beta-secretase processing of beta-amyloid precursor protein.J Biol Chem. 2010 Mar 19;285(12):8515-26. doi: 10.1074/jbc.M109.079079. Epub 2010 Jan 22. J Biol Chem. 2010. PMID: 20097758 Free PMC article.
-
Alzheimer's disease-associated mutations increase amyloid precursor protein resistance to γ-secretase cleavage and the Aβ42/Aβ40 ratio.Cell Discov. 2016 Aug 23;2:16026. doi: 10.1038/celldisc.2016.26. eCollection 2016. Cell Discov. 2016. PMID: 27625790 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

