Molecular epidemiology of bovine toroviruses circulating in South Korea
- PMID: 17719729
- PMCID: PMC7117412
- DOI: 10.1016/j.vetmic.2007.07.012
Molecular epidemiology of bovine toroviruses circulating in South Korea
Abstract
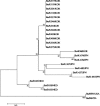
The prevalence of the bovine torovirus (BToV) and its genetic characterization have been reported in North America, Europe and Japan. Therefore, this study examined the prevalence and genetic diversity of the BToV in a total of 645 diarrheic fecal samples from 629 Korean native beef calf herds using RT-PCR and nested PCR with the primer pairs specific to a part of the BToV membrane (M) gene. Overall, 19 (2.9%) out of 645 diarrheic samples from 19 herds (6.9%) tested positive for BToVs by either RT-PCR or nested PCR. A comparison of the nucleotide (nt) and amino acid (aa) sequences of a part of the BToV M gene (409bp) among the BToVs showed the Korean BToVs to have comparatively higher sequence homology to the Japanese and Dutch BToVs than to the American and Italian BToVs. Generally, the Korean BToV strains clustered with the Japanese and Dutch BToV strains. However, the American and Italian BToV strains clustered on a separate major branch, suggesting that these are more distantly related to other known BToV strains. These results suggest that the BToV infections are sporadic in diarrheic calves in South Korea, and the Korean BToV strains are more closely related to the Japanese and Dutch BToVs than to the American and Italian BToVs.
Figures
Similar articles
-
Epidemiological analysis of bovine torovirus in Japan.Virus Res. 2007 Jun;126(1-2):32-7. doi: 10.1016/j.virusres.2007.01.013. Epub 2007 Feb 21. Virus Res. 2007. PMID: 17320234 Free PMC article.
-
First detection and molecular diversity of Brazilian bovine torovirus (BToV) strains from young and adult cattle.Res Vet Sci. 2013 Oct;95(2):799-801. doi: 10.1016/j.rvsc.2013.04.006. Epub 2013 May 3. Res Vet Sci. 2013. PMID: 23648077 Free PMC article.
-
Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events.J Virol. 2003 Sep;77(17):9567-77. doi: 10.1128/jvi.77.17.9567-9577.2003. J Virol. 2003. PMID: 12915570 Free PMC article.
-
Bovine torovirus (Breda virus) revisited.Anim Health Res Rev. 2004 Dec;5(2):157-71. doi: 10.1079/ahr200498. Anim Health Res Rev. 2004. PMID: 15984322 Review.
-
[A review of porcine torovirus research: etiology and epidemiology].Bing Du Xue Bao. 2013 Nov;29(6):667-72. Bing Du Xue Bao. 2013. PMID: 24520775 Review. Chinese.
Cited by
-
Development of universal SYBR Green real-time RT-PCR for the rapid detection and quantitation of bovine and porcine toroviruses.J Virol Methods. 2010 Sep;168(1-2):212-7. doi: 10.1016/j.jviromet.2010.06.001. Epub 2010 Jun 15. J Virol Methods. 2010. PMID: 20558206 Free PMC article.
-
Recent Progress in Torovirus Molecular Biology.Viruses. 2021 Mar 8;13(3):435. doi: 10.3390/v13030435. Viruses. 2021. PMID: 33800523 Free PMC article. Review.
-
Molecular characterization of a new PToV strain. Evolutionary implications.Virus Res. 2009 Jul;143(1):33-43. doi: 10.1016/j.virusres.2009.02.019. Epub 2009 Mar 20. Virus Res. 2009. PMID: 19463719 Free PMC article.
-
Characterization of Localization and Export Signals of Bovine Torovirus Nucleocapsid Protein Responsible for Extensive Nuclear and Nucleolar Accumulation and Their Importance for Virus Growth.J Virol. 2021 Jan 13;95(3):e02111-20. doi: 10.1128/JVI.02111-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33177195 Free PMC article.
-
Causative agents and epidemiology of diarrhea in Korean native calves.J Vet Sci. 2019 Nov;20(6):e64. doi: 10.4142/jvs.2019.20.e64. J Vet Sci. 2019. PMID: 31775191 Free PMC article.
References
-
- Ali A., Reynolds D.L. Characterization of the stunting syndrome agent: relatedness to known viruses. Avian Dis. 2000;44:45–50. - PubMed
-
- Asakura H., Makino S., Shirahata T., Tsukamoto T., Kurazono H., Ikeda T., Takeshi K. Detection and genetical characterization of Shiga toxin-producing Escherichia coli from wild deer. Microbiol. Immunol. 1998;42:815–822. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials