Mechanism of the CO-sensing heme protein CooA: new insights from the truncated heme domain and UVRR spectroscopy
- PMID: 17720248
- PMCID: PMC2096632
- DOI: 10.1016/j.jinorgbio.2007.07.010
Mechanism of the CO-sensing heme protein CooA: new insights from the truncated heme domain and UVRR spectroscopy
Abstract
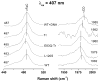
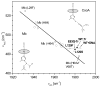
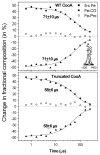
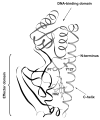
The bacterial CO-sensing heme protein CooA activates expression of genes whose products perform CO-metabolism by binding its target DNA in response to CO binding. The required conformational change has been proposed to result from CO-induced displacement of the heme and of the adjacent C-helix, which connects the sensory and DNA-binding domains. Support for this proposal comes from UV Resonance Raman (UVRR) spectroscopy, which reveals a more hydrophobic environment for the C-helix residue Trp110 when CO binds. In addition, we find a tyrosine UVRR response, which is attributable to weakening of a Tyr55-Glu83 H-bond that anchors the proximal side of the heme. Both Trp and Tyr responses are augmented in the heme domain when the DNA-binding domain has been removed, apparently reflecting loss of the inter-domain restraint. This augmentation is abolished by a Glu83Gln substitution, which weakens the anchoring H-bond. The CO recombination rate following photolysis of the CO adduct is similar for truncated and full-length protein, though truncation does increase the rate of CO association in the absence of photolysis; together these data indicate that truncation causes a faster dissociation of the endogenous Pro2 ligand. These findings are discussed in the light of structural evidence that the N-terminal tail, once released from the heme, selects the proper orientation of the DNA-binding domain, via docking interactions.
Figures




Similar articles
-
Evidence for displacements of the C-helix by CO ligation and DNA binding to CooA revealed by UV resonance Raman spectroscopy.J Biol Chem. 2006 Apr 21;281(16):11271-8. doi: 10.1074/jbc.M513261200. Epub 2006 Jan 26. J Biol Chem. 2006. PMID: 16439368
-
Activation mechanism of the CO sensor CooA. Mutational and resonance Raman spectroscopic studies.J Biol Chem. 2003 Sep 12;278(37):35384-93. doi: 10.1074/jbc.M301000200. Epub 2003 Jun 9. J Biol Chem. 2003. PMID: 12796503
-
Heme displacement mechanism of CooA activation: mutational and Raman spectroscopic evidence.J Biol Chem. 2006 Sep 29;281(39):29165-73. doi: 10.1074/jbc.M605568200. Epub 2006 Jul 26. J Biol Chem. 2006. PMID: 16873369 Free PMC article.
-
CooA, a paradigm for gas sensing regulatory proteins.J Inorg Biochem. 2005 Jan;99(1):280-92. doi: 10.1016/j.jinorgbio.2004.10.032. J Inorg Biochem. 2005. PMID: 15598507 Review.
-
Biochemical and biophysical properties of the CO-sensing transcriptional activator CooA.Acc Chem Res. 2003 Nov;36(11):825-31. doi: 10.1021/ar020097p. Acc Chem Res. 2003. PMID: 14622029 Review.
Cited by
-
Regulation of multiple carbon monoxide consumption pathways in anaerobic bacteria.Front Microbiol. 2011 Jul 11;2:147. doi: 10.3389/fmicb.2011.00147. eCollection 2011. Front Microbiol. 2011. PMID: 21808633 Free PMC article.
-
The diagnostic vibrational signature of pentacoordination in heme carbonyls.J Am Chem Soc. 2014 Jul 16;136(28):9818-21. doi: 10.1021/ja503191z. Epub 2014 Jul 7. J Am Chem Soc. 2014. PMID: 24950373 Free PMC article.
-
Interactions of multiple gas-transducing systems: hallmarks and uncertainties of CO, NO, and H2S gas biology.Antioxid Redox Signal. 2010 Jul 15;13(2):157-92. doi: 10.1089/ars.2009.2657. Antioxid Redox Signal. 2010. PMID: 19939208 Free PMC article. Review.
-
Early processes in heme-based CO-sensing proteins.Front Mol Biosci. 2022 Nov 3;9:1046412. doi: 10.3389/fmolb.2022.1046412. eCollection 2022. Front Mol Biosci. 2022. PMID: 36406263 Free PMC article. Review.
-
Carbon monoxide: impact on remethylation/transsulfuration metabolism and its pathophysiologic implications.J Mol Med (Berl). 2012 Mar;90(3):245-54. doi: 10.1007/s00109-012-0875-2. Epub 2012 Feb 14. J Mol Med (Berl). 2012. PMID: 22331189 Free PMC article. Review.
References
-
- Gilles-Gonzalez MA, Gonzalez G. J Inorg Biochem. 2005;99:1–22. - PubMed
-
- Roberts GP, Kerby RL, Youn H, Conrad M. J Inorg Biochem. 2005;99:280–292. - PubMed
-
- Aono S. Acc Chem Res. 2003;36:825–831. - PubMed
-
- Kerby RL, Youn H, Thorsteinsson MV, Roberts GP. J Mol Biol. 2003;325:809–823. - PubMed
-
- Youn H, Kerby RL, Roberts GP. J Biol Chem. 2003;278:2333–2340. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

