CTnDOT integrase performs ordered homology-dependent and homology-independent strand exchanges
- PMID: 17720706
- PMCID: PMC2034462
- DOI: 10.1093/nar/gkm637
CTnDOT integrase performs ordered homology-dependent and homology-independent strand exchanges
Abstract
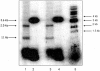
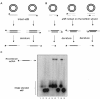
Although the integrase (IntDOT) of the Bacteroides conjugative transposon CTnDOT has been classified as a member of the tyrosine recombinase family, the reaction it catalyzes appears to differ in some features from reactions catalyzed by other tyrosine recombinases. We tested the ability of IntDOT to cleave and ligate activated attDOT substrates in the presence of mismatches. Unlike other tyrosine recombinases, the results revealed that IntDOT is able to perform ligation reactions even when all the bases within the crossover region are mispaired. We also show that there is a strong bias in the order of strand exchanges during integrative recombination. The top strands are exchanged first in reactions that appear to require 2 bp of homology between the partner sites adjacent to the sites of cleavage. The bottom strands are exchanged next in reactions that do not require homology between the partner sites. This mode of coordination of strand exchanges is unique among tyrosine recombinases.
Figures







Similar articles
-
The Integration and Excision of CTnDOT.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0020-2014. doi: 10.1128/microbiolspec.MDNA3-0020-2014. Microbiol Spectr. 2015. PMID: 26104696 Free PMC article. Review.
-
Resolution of Mismatched Overlap Holliday Junction Intermediates by the Tyrosine Recombinase IntDOT.J Bacteriol. 2017 Apr 25;199(10):e00873-16. doi: 10.1128/JB.00873-16. Print 2017 May 15. J Bacteriol. 2017. PMID: 28242723 Free PMC article.
-
Challenging a paradigm: the role of DNA homology in tyrosine recombinase reactions.Microbiol Mol Biol Rev. 2009 Jun;73(2):300-9. doi: 10.1128/MMBR.00038-08. Microbiol Mol Biol Rev. 2009. PMID: 19487729 Free PMC article. Review.
-
Characterization of a conjugative transposon integrase, IntDOT.Mol Microbiol. 2006 Jun;60(5):1228-40. doi: 10.1111/j.1365-2958.2006.05164.x. Mol Microbiol. 2006. PMID: 16689798
-
The N-terminus of IntDOT forms hydrophobic interactions during Holliday Junction resolution.Plasmid. 2016 Sep-Nov;87-88:10-16. doi: 10.1016/j.plasmid.2016.07.003. Epub 2016 Jul 12. Plasmid. 2016. PMID: 27422335 Free PMC article.
Cited by
-
Resolution of Holliday junction recombination intermediates by wild-type and mutant IntDOT proteins.J Bacteriol. 2011 Mar;193(6):1351-8. doi: 10.1128/JB.01465-10. Epub 2011 Jan 7. J Bacteriol. 2011. PMID: 21216992 Free PMC article.
-
Molecular keys of the tropism of integration of the cholera toxin phage.Proc Natl Acad Sci U S A. 2010 Mar 2;107(9):4377-82. doi: 10.1073/pnas.0910212107. Epub 2010 Jan 21. Proc Natl Acad Sci U S A. 2010. PMID: 20133778 Free PMC article.
-
The Integration and Excision of CTnDOT.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0020-2014. doi: 10.1128/microbiolspec.MDNA3-0020-2014. Microbiol Spectr. 2015. PMID: 26104696 Free PMC article. Review.
-
Resolution of Mismatched Overlap Holliday Junction Intermediates by the Tyrosine Recombinase IntDOT.J Bacteriol. 2017 Apr 25;199(10):e00873-16. doi: 10.1128/JB.00873-16. Print 2017 May 15. J Bacteriol. 2017. PMID: 28242723 Free PMC article.
-
Challenging a paradigm: the role of DNA homology in tyrosine recombinase reactions.Microbiol Mol Biol Rev. 2009 Jun;73(2):300-9. doi: 10.1128/MMBR.00038-08. Microbiol Mol Biol Rev. 2009. PMID: 19487729 Free PMC article. Review.
References
-
- Salyers AA, Shipman JA. 11: Getting in touch with your prokaryotic self: mammal-microbe interactions. In: Staley JT, Reysenbach A.-L, editors. Biodiversity of Microbial Life. Wiley-Liss, Inc.; 2002. pp. 315–341.
-
- Macy JM, Probst I. The biology of gastrointestinal bacteroides. Annu. Rev. Microbiol. 1979;33:561–594. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

