The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates
- PMID: 17720711
- PMCID: PMC2034482
- DOI: 10.1093/nar/gkm613
The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates
Abstract
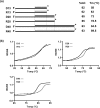
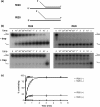
Pif1p is the prototypical member of the PIF1 family of DNA helicases, a subfamily of SFI helicases conserved from yeast to humans. Baker's yeast Pif1p is involved in the maintenance of mitochondrial, ribosomal and telomeric DNA and may also have a general role in chromosomal replication by affecting Okazaki fragment maturation. Here we investigate the substrate preferences for Pif1p. The enzyme was preferentially active on RNA-DNA hybrids, as seen by faster unwinding rates on RNA-DNA hybrids compared to DNA-DNA hybrids. When using forked substrates, which have been shown previously to stimulate the enzyme, Pif1p demonstrated a preference for RNA-DNA hybrids. This preferential unwinding could not be correlated to preferential binding of Pif1p to the substrates that were the most readily unwound. Although the addition of the single-strand DNA-binding protein replication protein A (RPA) stimulated the helicase reaction on all substrates, it did not diminish the preference of Pif1p for RNA-DNA substrates. Thus, forked RNA-DNA substrates are the favored substrates for Pif1p in vitro. We discuss these findings in terms of the known biological roles of the enzyme.
Figures






References
-
- Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: integration of helicases into cellular processes. Q. Rev. Biophys. 2003;36:1–69. - PubMed
-
- Bessler JB, Torres JZ, Zakian VA. The Pif1p subfamily of helicases: region-specific DNA helicases? Trends Cell. Biol. 2001;11:60–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

