Analysis of the XPA and ssDNA-binding surfaces on the central domain of human ERCC1 reveals evidence for subfunctionalization
- PMID: 17720715
- PMCID: PMC2034474
- DOI: 10.1093/nar/gkm503
Analysis of the XPA and ssDNA-binding surfaces on the central domain of human ERCC1 reveals evidence for subfunctionalization
Abstract
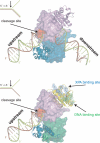
Human ERCC1/XPF is a structure-specific endonuclease involved in multiple DNA repair pathways. We present the solution structure of the non-catalytic ERCC1 central domain. Although this domain shows structural homology with the catalytically active XPF nuclease domain, functional investigation reveals a completely distinct function for the ERCC1 central domain by performing interactions with both XPA and single-stranded DNA. These interactions are non-competitive and can occur simultaneously through distinct interaction surfaces. Interestingly, the XPA binding by ERCC1 and the catalytic function of XPF are dependent on a structurally homologous region of the two proteins. Although these regions are strictly conserved in each protein family, amino acid composition and surface characteristics are distinct. We discuss the possibility that after XPF gene duplication, the redundant ERCC1 central domain acquired novel functions, thereby increasing the fidelity of eukaryotic DNA repair.
Figures





References
-
- Nishino T, Morikawa K. Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors. Oncogene. 2002;21:9022–9032. - PubMed
-
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. - PubMed
-
- Nishino T, Komori K, Ishino Y, Morikawa K. X-ray and biochemical anatomy of an archaeal XPF/Rad1/Mus81 family nuclease: similarity between its endonuclease domain and restriction enzymes. Structure (Camb.) 2003;11:445–457. - PubMed
-
- Heyer WD, Ehmsen KT, Solinger JA. Holliday junctions in the eukaryotic nucleus: resolution in sight? Trends Biochem. Sci. 2003;28:548–557. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

