The dynamic orientation of membrane-bound peptides: bridging simulations and experiments
- PMID: 17720729
- PMCID: PMC2098706
- DOI: 10.1529/biophysj.107.113043
The dynamic orientation of membrane-bound peptides: bridging simulations and experiments
Abstract
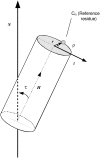
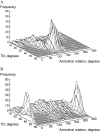
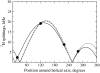
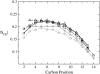
The structural organization in a peptide/membrane supramolecular complex is best described by knowledge of the peptide orientation plus its time-dependent and spatial fluctuations. The static orientation, defined by the peptide tilt and a rotation about its molecular axis, is accessible through a number of spectroscopic methods. However, peptide dynamics, although relevant to understand the functionality of these systems, remains largely unexplored. Here, we describe the orientation and dynamics of Trp-flanked and Lys-flanked hydrophobic peptides in a lipid bilayer from molecular dynamics simulations. A novel view is revealed, where collective nontrivial distributions of time-evolving and ensemble peptide orientations closely represent the systems as studied experimentally. Such global distributions are broad and unveil the existence of orientational states, which depend on the anchoring mode of interfacial residues. We show that this dynamics modulates (2)H quadrupolar splittings and introduces ambiguity in the analysis of NMR data. These findings demonstrate that structural descriptions of peptide/membrane complexes are incomplete, and in cases even imprecise, without knowledge of dynamics.
Figures

References
-
- Singer, S. J., and G. L. Nicolson. 1972. The fluid mosaic model of the structure of cell membranes. Science. 175:720–731. - PubMed
-
- de Planque, M. R., E. Goormaghtigh, D. V. Greathouse, R. E. Koeppe 2nd, J. A. Kruijtzer, R. M. Liskamp, B. de Kruijff, and J. A. Killian. 2001. Sensitivity of single membrane-spanning α-helical peptides to hydrophobic mismatch with a lipid bilayer: effects on backbone structure, orientation, and extent of membrane incorporation. Biochemistry. 40:5000–5010. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

