TcpA, an FtsK/SpoIIIE homolog, is essential for transfer of the conjugative plasmid pCW3 in Clostridium perfringens
- PMID: 17720795
- PMCID: PMC2168741
- DOI: 10.1128/JB.00783-07
TcpA, an FtsK/SpoIIIE homolog, is essential for transfer of the conjugative plasmid pCW3 in Clostridium perfringens
Abstract
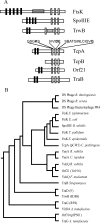
The conjugative tetracycline resistance plasmid pCW3 is the paradigm conjugative plasmid in the anaerobic gram-positive pathogen Clostridium perfringens. Two closely related FtsK/SpoIIIE homologs, TcpA and TcpB, are encoded on pCW3, which is significant since FtsK domains are found in coupling proteins of gram-negative conjugation systems. To develop an understanding of the mechanism of conjugative transfer in C. perfringens, we determined the role of these proteins in the conjugation process. Mutation and complementation analysis was used to show that the tcpA gene was essential for the conjugative transfer of pCW3 and that the tcpB gene was not required for transfer. Furthermore, complementation of a pCW3DeltatcpA mutant with divergent tcpA homologs provided experimental evidence that all of the known conjugative plasmids from C. perfringens use a similar transfer mechanism. Functional genetic analysis of the TcpA protein established the essential role in conjugative transfer of its Walker A and Walker B ATP-binding motifs and its FtsK-like RAAG motif. It is postulated that TcpA is the essential DNA translocase or coupling protein encoded by pCW3 and as such represents a key component of the unique conjugation process in C. perfringens.
Figures

Similar articles
-
TcpA from the Clostridium perfringens plasmid pCW3 is more closely related to the DNA translocase FtsK than to coupling proteins.Structure. 2023 Apr 6;31(4):455-463.e4. doi: 10.1016/j.str.2023.02.001. Epub 2023 Feb 24. Structure. 2023. PMID: 36841236
-
Two novel membrane proteins, TcpD and TcpE, are essential for conjugative transfer of pCW3 in Clostridium perfringens.J Bacteriol. 2015 Feb 15;197(4):774-81. doi: 10.1128/JB.02466-14. Epub 2014 Dec 8. J Bacteriol. 2015. PMID: 25488300 Free PMC article.
-
Functional identification of conjugation and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens.J Bacteriol. 2006 Jul;188(13):4942-51. doi: 10.1128/JB.00298-06. J Bacteriol. 2006. PMID: 16788202 Free PMC article.
-
Antibiotic resistance plasmids and mobile genetic elements of Clostridium perfringens.Plasmid. 2018 Sep;99:32-39. doi: 10.1016/j.plasmid.2018.07.002. Epub 2018 Jul 26. Plasmid. 2018. PMID: 30055188 Review.
-
The Tcp conjugation system of Clostridium perfringens.Plasmid. 2017 May;91:28-36. doi: 10.1016/j.plasmid.2017.03.001. Epub 2017 Mar 7. Plasmid. 2017. PMID: 28286218 Review.
Cited by
-
BtpB, a novel Brucella TIR-containing effector protein with immune modulatory functions.Front Cell Infect Microbiol. 2013 Jul 8;3:28. doi: 10.3389/fcimb.2013.00028. eCollection 2013. Front Cell Infect Microbiol. 2013. PMID: 23847770 Free PMC article.
-
The transjugation machinery of Thermus thermophilus: Identification of TdtA, an ATPase involved in DNA donation.PLoS Genet. 2017 Mar 10;13(3):e1006669. doi: 10.1371/journal.pgen.1006669. eCollection 2017 Mar. PLoS Genet. 2017. PMID: 28282376 Free PMC article.
-
Evolution of conjugation and type IV secretion systems.Mol Biol Evol. 2013 Feb;30(2):315-31. doi: 10.1093/molbev/mss221. Epub 2012 Sep 13. Mol Biol Evol. 2013. PMID: 22977114 Free PMC article.
-
Toxin plasmids of Clostridium perfringens.Microbiol Mol Biol Rev. 2013 Jun;77(2):208-33. doi: 10.1128/MMBR.00062-12. Microbiol Mol Biol Rev. 2013. PMID: 23699255 Free PMC article. Review.
-
Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.Nat Commun. 2018 Sep 13;9(1):3732. doi: 10.1038/s41467-018-06096-2. Nat Commun. 2018. PMID: 30213934 Free PMC article.
References
-
- Abraham, L. J., and J. I. Rood. 1985. Cloning and analysis of the Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid 13:155-162. - PubMed
-
- Abraham, L. J., A. J. Wales, and J. I. Rood. 1985. Worldwide distribution of the conjugative Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid 14:37-46. - PubMed
-
- Aussel, L., F.-X. Barre, M. Aroyo, A. Stasiak, A. Z. Stasiak, and D. Sherratt. 2002. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108:195-205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

