Comparison of ergot alkaloid biosynthesis gene clusters in Claviceps species indicates loss of late pathway steps in evolution of C. fusiformis
- PMID: 17720822
- PMCID: PMC2168186
- DOI: 10.1128/AEM.01040-07
Comparison of ergot alkaloid biosynthesis gene clusters in Claviceps species indicates loss of late pathway steps in evolution of C. fusiformis
Abstract
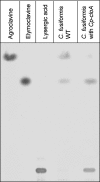
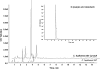
The grass parasites Claviceps purpurea and Claviceps fusiformis produce ergot alkaloids (EA) in planta and in submerged culture. Whereas EA synthesis (EAS) in C. purpurea proceeds via clavine intermediates to lysergic acid and the complex ergopeptines, C. fusiformis produces only agroclavine and elymoclavine. In C. purpurea the EAS gene (EAS) cluster includes dmaW (encoding the first pathway step), cloA (elymoclavine oxidation to lysergic acid), and the lpsA/lpsB genes (ergopeptine formation). We analyzed the corresponding C. fusiformis EAS cluster to investigate the evolutionary basis for chemotypic differences between the Claviceps species. Other than three peptide synthetase genes (lpsC and the tandem paralogues lpsA1 and lpsA2), homologues of all C. purpurea EAS genes were identified in C. fusiformis, including homologues of lpsB and cloA, which in C. purpurea encode enzymes for steps after clavine synthesis. Rearrangement of the cluster was evident around lpsB, which is truncated in C. fusiformis. This and several frameshift mutations render CflpsB a pseudogene (CflpsB(Psi)). No obvious inactivating mutation was identified in CfcloA. All C. fusiformis EAS genes, including CflpsB(Psi) and CfcloA, were expressed in culture. Cross-complementation analyses demonstrated that CfcloA and CflpsB(Psi) were expressed in C. purpurea but did not encode functional enzymes. In contrast, CpcloA catalyzed lysergic acid biosynthesis in C. fusiformis, indicating that C. fusiformis terminates its EAS pathway at elymoclavine because the cloA gene product is inactive. We propose that the C. fusiformis EAS cluster evolved from a more complete cluster by loss of some lps genes and by rearrangements and mutations inactivating lpsB and cloA.
Figures
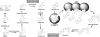
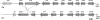




Similar articles
-
Identification of the cytochrome P450 monooxygenase that bridges the clavine and ergoline alkaloid pathways.Chembiochem. 2006 Apr;7(4):645-52. doi: 10.1002/cbic.200500487. Chembiochem. 2006. PMID: 16538694
-
The ergot alkaloid gene cluster: functional analyses and evolutionary aspects.Phytochemistry. 2009 Oct-Nov;70(15-16):1822-32. doi: 10.1016/j.phytochem.2009.05.023. Epub 2009 Aug 18. Phytochemistry. 2009. PMID: 19695648 Review.
-
Ergot alkaloids--biology and molecular biology.Alkaloids Chem Biol. 2006;63:45-86. doi: 10.1016/s1099-4831(06)63002-2. Alkaloids Chem Biol. 2006. PMID: 17133714 Review.
-
The ergot alkaloid gene cluster in Claviceps purpurea: extension of the cluster sequence and intra species evolution.Phytochemistry. 2005 Jun;66(11):1312-20. doi: 10.1016/j.phytochem.2005.04.011. Phytochemistry. 2005. PMID: 15904941
-
Evidence for an ergot alkaloid gene cluster in Claviceps purpurea.Mol Gen Genet. 1999 Feb;261(1):133-41. doi: 10.1007/s004380050950. Mol Gen Genet. 1999. PMID: 10071219
Cited by
-
Chromosome rearrangements shape the diversification of secondary metabolism in the cyclosporin producing fungus Tolypocladium inflatum.BMC Genomics. 2019 Feb 7;20(1):120. doi: 10.1186/s12864-018-5399-x. BMC Genomics. 2019. PMID: 30732559 Free PMC article.
-
Crystallization and preliminary X-ray analysis of the ergothioneine-biosynthetic methyltransferase EgtD.Acta Crystallogr F Struct Biol Commun. 2014 May;70(Pt 5):676-80. doi: 10.1107/S2053230X1400805X. Epub 2014 Apr 25. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 24817736 Free PMC article.
-
Ergot Alkaloids of the Family Clavicipitaceae.Phytopathology. 2017 May;107(5):504-518. doi: 10.1094/PHYTO-12-16-0435-RVW. Epub 2017 Mar 29. Phytopathology. 2017. PMID: 28168931 Free PMC article. Review.
-
Plant-symbiotic fungi as chemical engineers: multi-genome analysis of the clavicipitaceae reveals dynamics of alkaloid loci.PLoS Genet. 2013;9(2):e1003323. doi: 10.1371/journal.pgen.1003323. Epub 2013 Feb 28. PLoS Genet. 2013. PMID: 23468653 Free PMC article.
-
Secondary metabolite biosynthetic diversity in the fungal family Hypoxylaceae and Xylaria hypoxylon.Stud Mycol. 2021 Aug 26;99:100118. doi: 10.1016/j.simyco.2021.100118. eCollection 2021 Jun. Stud Mycol. 2021. PMID: 34527085 Free PMC article.
References
-
- Berde, B., and E. Stürmer. 1978. Introduction to the pharmacology of ergot alkaloids and related compounds, p. 1-28. In W. H. Aellig, B. Berde, and H. O. Schild (ed.), Ergot alkaloids and related compounds. Springer, Berlin, Germany.
-
- Bové, F. J. 1970. The story of ergot. S. Karger, Basel, Switzerland.
-
- Correia, T., N. Grammel, I. Ortel, U. Keller, and P. Tudzynski. 2003. Molecular cloning and analysis of the ergopeptine assembly system in the ergot fungus Claviceps purpurea. Chem. Biol. 10:1281-1292. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

