Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum
- PMID: 17720833
- PMCID: PMC2075035
- DOI: 10.1128/AEM.01425-07
Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum
Abstract
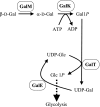
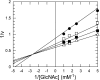
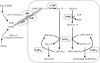
We have determined the functions of the enzymes encoded by the lnpB, lnpC, and lnpD genes, located downstream of the lacto-N-biose phosphorylase gene (lnpA), in Bifidobacterium longum JCM1217. The lnpB gene encodes a novel kinase, N-acetylhexosamine 1-kinase, which produces N-acetylhexosamine 1-phosphate; the lnpC gene encodes UDP-glucose hexose 1-phosphate uridylyltransferase, which is also active on N-acetylhexosamine 1-phosphate; and the lnpD gene encodes a UDP-glucose 4-epimerase, which is active on both UDP-galactose and UDP-N-acetylgalactosamine. These results suggest that the gene operon lnpABCD encodes a previously undescribed lacto-N-biose I/galacto-N-biose metabolic pathway that is involved in the intestinal colonization of bifidobacteria and that utilizes lacto-N-biose I from human milk oligosaccharides or galacto-N-biose from mucin sugars.
Figures
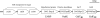
Similar articles
-
Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides.Adv Nutr. 2012 May 1;3(3):422S-9S. doi: 10.3945/an.111.001420. Adv Nutr. 2012. PMID: 22585921 Free PMC article. Review.
-
Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum.Appl Environ Microbiol. 2005 Jun;71(6):3158-62. doi: 10.1128/AEM.71.6.3158-3162.2005. Appl Environ Microbiol. 2005. PMID: 15933016 Free PMC article.
-
Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.Acta Crystallogr D Biol Crystallogr. 2014 May;70(Pt 5):1401-10. doi: 10.1107/S1399004714004209. Epub 2014 Apr 30. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 24816108
-
Identification of lacto-N-Biose I phosphorylase from Vibrio vulnificus CMCP6.Appl Environ Microbiol. 2008 Oct;74(20):6333-7. doi: 10.1128/AEM.02846-07. Epub 2008 Aug 22. Appl Environ Microbiol. 2008. PMID: 18723650 Free PMC article.
-
Unique sugar metabolic pathways of bifidobacteria.Biosci Biotechnol Biochem. 2010;74(12):2374-84. doi: 10.1271/bbb.100494. Epub 2010 Dec 7. Biosci Biotechnol Biochem. 2010. PMID: 21150123 Review.
Cited by
-
Mixed-species genomic microarray analysis of fecal samples reveals differential transcriptional responses of bifidobacteria in breast- and formula-fed infants.Appl Environ Microbiol. 2009 May;75(9):2668-76. doi: 10.1128/AEM.02492-08. Epub 2009 Mar 13. Appl Environ Microbiol. 2009. PMID: 19286790 Free PMC article.
-
Effect of Lacto-N-biose I on the Antigen-specific Immune Responses of Splenocytes.Biosci Microbiota Food Health. 2012;31(2):47-50. doi: 10.12938/bmfh.31.47. Epub 2012 Apr 20. Biosci Microbiota Food Health. 2012. PMID: 24936348 Free PMC article.
-
Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem.ISME J. 2020 Feb;14(2):635-648. doi: 10.1038/s41396-019-0553-2. Epub 2019 Nov 18. ISME J. 2020. PMID: 31740752 Free PMC article.
-
Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides.Adv Nutr. 2012 May 1;3(3):422S-9S. doi: 10.3945/an.111.001420. Adv Nutr. 2012. PMID: 22585921 Free PMC article. Review.
-
Proteinaceous Molecules Mediating Bifidobacterium-Host Interactions.Front Microbiol. 2016 Aug 3;7:1193. doi: 10.3389/fmicb.2016.01193. eCollection 2016. Front Microbiol. 2016. PMID: 27536282 Free PMC article. Review.
References
-
- Ajdic, D., I. C. Sutcliffe, R. R. Russell, and J. J. Ferretti. 1996. Organization and nucleotide sequence of the Streptococcus mutans galactose operon. Gene 180:137-144. - PubMed
-
- Benno, Y., and T. Mitsuoka. 1986. The development of gastrointestinal micro-flora in humans and animals. Bifidobact. Microflora 5:13-25.
-
- Benno, Y., K. Sawada, and T. Mitsuoka. 1984. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol. Immunol. 28:975-986. - PubMed
-
- Bouffard, G. G., K. E. Rudd, and S. L. Adhya. 1994. Dependence of lactose metabolism upon mutarotase encoded in the gal operon in Escherichia coli. J. Mol. Biol. 244:269-278. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

