Genetic screen for regulatory mutations in Methanococcus maripaludis and its use in identification of induction-deficient mutants of the euryarchaeal repressor NrpR
- PMID: 17720835
- PMCID: PMC2075042
- DOI: 10.1128/AEM.01324-07
Genetic screen for regulatory mutations in Methanococcus maripaludis and its use in identification of induction-deficient mutants of the euryarchaeal repressor NrpR
Abstract
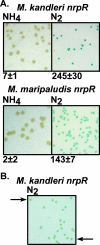
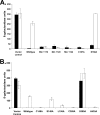
NrpR is an euryarchaeal transcriptional repressor of nitrogen assimilation genes. Previous studies with Methanococcus maripaludis demonstrated that NrpR binds to palindromic operator sequences, blocking transcription initiation. The metabolite 2-oxoglutarate, an indicator of cellular nitrogen deficiency, induces transcription by lowering the affinity of NrpR for operator DNA. In this report we build on existing genetic tools for M. maripaludis to develop a screen for change-of-function mutations in a transcriptional regulator and demonstrate the use of an X-Gal (5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside) screen for strict anaerobes. We use the approach to address the primary structural requirements for the response of NrpR to 2-oxoglutarate. nrpR genes from the mesophilic M. maripaludis and the hyperthermophilic Methanopyrus kandleri were targeted for mutagenesis. M. maripaludis nrpR encodes a protein with two homologous NrpR domains while the M. kandleri nrpR homolog encodes a single NrpR domain. Random point mutagenesis and alanine replacement mutagenesis identified two amino acid residues of M. kandleri NrpR involved in induction of gene expression under nitrogen-deficient conditions and thus in the response to 2-oxoglutarate. Mutagenesis of the corresponding regions in either domain of M. maripaludis NrpR resulted in a similar effect, demonstrating a conserved structure-function relationship between the two repressors. The results indicate that in M. maripaludis, both NrpR domains participate in the 2-oxoglutarate response. The approach used here has wide adaptability to other regulatory systems in methanogenic Archaea and other strict anaerobes.
Figures

Similar articles
-
Metabolic processes of Methanococcus maripaludis and potential applications.Microb Cell Fact. 2016 Jun 10;15(1):107. doi: 10.1186/s12934-016-0500-0. Microb Cell Fact. 2016. PMID: 27286964 Free PMC article. Review.
-
Regulation of nif expression in Methanococcus maripaludis: roles of the euryarchaeal repressor NrpR, 2-oxoglutarate, and two operators.J Biol Chem. 2005 Feb 18;280(7):5236-41. doi: 10.1074/jbc.M411778200. Epub 2004 Dec 7. J Biol Chem. 2005. PMID: 15590692
-
Overlapping repressor binding sites regulate expression of the Methanococcus maripaludis glnK(1) operon.Mol Microbiol. 2010 Feb;75(3):755-62. doi: 10.1111/j.1365-2958.2009.07016.x. Epub 2009 Dec 16. Mol Microbiol. 2010. PMID: 20025661 Free PMC article.
-
A novel repressor of nif and glnA expression in the methanogenic archaeon Methanococcus maripaludis.Mol Microbiol. 2003 Jan;47(1):235-46. doi: 10.1046/j.1365-2958.2003.03293.x. Mol Microbiol. 2003. PMID: 12492867
-
Methanococcus maripaludis: an archaeon with multiple functional MCM proteins?Biochem Soc Trans. 2009 Feb;37(Pt 1):1-6. doi: 10.1042/BST0370001. Biochem Soc Trans. 2009. PMID: 19143592 Review.
Cited by
-
Structural underpinnings of nitrogen regulation by the prototypical nitrogen-responsive transcriptional factor NrpR.Structure. 2010 Nov 10;18(11):1512-21. doi: 10.1016/j.str.2010.08.014. Structure. 2010. PMID: 21070950 Free PMC article.
-
Scale-up of biomass production by Methanococcus maripaludis.Front Microbiol. 2022 Nov 23;13:1031131. doi: 10.3389/fmicb.2022.1031131. eCollection 2022. Front Microbiol. 2022. PMID: 36504798 Free PMC article.
-
Tools, Strains, and Strategies To Effectively Conduct Anaerobic and Aerobic Transcriptional Reporter Screens and Assays in Staphylococcus aureus.Appl Environ Microbiol. 2021 Oct 14;87(21):e0110821. doi: 10.1128/AEM.01108-21. Epub 2021 Aug 18. Appl Environ Microbiol. 2021. PMID: 34406831 Free PMC article.
-
Small RNAs Involved in Regulation of Nitrogen Metabolism.Microbiol Spectr. 2018 Jul;6(4):10.1128/microbiolspec.rwr-0018-2018. doi: 10.1128/microbiolspec.RWR-0018-2018. Microbiol Spectr. 2018. PMID: 30027888 Free PMC article. Review.
-
Metabolic processes of Methanococcus maripaludis and potential applications.Microb Cell Fact. 2016 Jun 10;15(1):107. doi: 10.1186/s12934-016-0500-0. Microb Cell Fact. 2016. PMID: 27286964 Free PMC article. Review.
References
-
- Ehlers, C., K. Weidenbach, K. Veit, K. Forchhammer, and R. A. Schmitz. 2005. Unique mechanistic features of post-translational regulation of glutamine synthetase activity in Methanosarcina mazei strain Go1 in response to nitrogen availability. Mol. Microbiol. 55:1841-1854. - PubMed
-
- Forchhammer, K. 2007. Glutamine signalling in bacteria. Front. Biosci. 12:358-370. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

