Low-pH adaptation and the acid tolerance response of Bifidobacterium longum biotype longum
- PMID: 17720838
- PMCID: PMC2075061
- DOI: 10.1128/AEM.00886-07
Low-pH adaptation and the acid tolerance response of Bifidobacterium longum biotype longum
Abstract
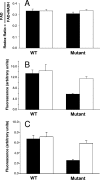
Bifidobacteria are one of the main microbial inhabitants of the human colon. Usually administered in fermented dairy products as beneficial microorganisms, they have to overcome the acidic pH found in the stomach during the gastrointestinal transit to be able to colonize the lower parts of the intestine. The mechanisms underlying acid response and adaptation in Bifidobacterium longum biotype longum NCIMB 8809 and its acid-pH-resistant mutant B. longum biotype longum 8809dpH were studied. Comparison of protein maps, and protein identification by matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis, allowed us to identify nine different proteins whose production largely changed in the mutant strain. Furthermore, the production of 47 proteins was modulated by pH in one or both strains. These included general stress response chaperones and proteins involved in transcription and translation as well as in carbohydrate and nitrogen metabolism, among others. Significant differences in the levels of metabolic end products and in the redox status of the cells were also detected between the wild-type strain and its acid-pH-resistant mutant in response to, or as a result of, adaptation to acid. Remarkably, the results of this work indicated that adaptation and response to low pH in B. longum biotype longum involve changes in the glycolytic flux and in the ability to regulate the internal pH. These changes were accompanied by a higher content of ammonium in the cytoplasm, likely coming from amino acid deamination, and a decrease of the bile salt hydrolase activity.
Figures



Similar articles
-
Analysis of host-inducing proteome changes in bifidobacterium longum NCC2705 grown in Vivo.J Proteome Res. 2008 Jan;7(1):375-85. doi: 10.1021/pr0704940. Epub 2007 Nov 21. J Proteome Res. 2008. PMID: 18027903
-
Global transcriptome analysis of the heat shock response of Bifidobacterium longum.FEMS Microbiol Lett. 2007 Jun;271(1):136-45. doi: 10.1111/j.1574-6968.2007.00704.x. Epub 2007 Apr 10. FEMS Microbiol Lett. 2007. PMID: 17419761
-
Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809.J Bacteriol. 2005 Aug;187(16):5799-808. doi: 10.1128/JB.187.16.5799-5808.2005. J Bacteriol. 2005. PMID: 16077128 Free PMC article.
-
Bosom Buddies: The Symbiotic Relationship Between Infants and Bifidobacterium longum ssp. longum and ssp. infantis. Genetic and Probiotic Features.Annu Rev Food Sci Technol. 2016;7:1-21. doi: 10.1146/annurev-food-041715-033151. Annu Rev Food Sci Technol. 2016. PMID: 26934170 Review.
-
How high G+C Gram-positive bacteria and in particular bifidobacteria cope with heat stress: protein players and regulators.FEMS Microbiol Rev. 2006 Sep;30(5):734-59. doi: 10.1111/j.1574-6976.2006.00031.x. FEMS Microbiol Rev. 2006. PMID: 16911042 Review.
Cited by
-
A bile-inducible membrane protein mediates bifidobacterial bile resistance.Microb Biotechnol. 2012 Jul;5(4):523-35. doi: 10.1111/j.1751-7915.2011.00329.x. Epub 2012 Feb 1. Microb Biotechnol. 2012. PMID: 22296641 Free PMC article.
-
HspR mutations are naturally selected in Bifidobacterium longum when successive heat shock treatments are applied.J Bacteriol. 2010 Jan;192(1):256-63. doi: 10.1128/JB.01147-09. J Bacteriol. 2010. PMID: 19880603 Free PMC article.
-
Analysis of Lactobacillus sakei mutants selected after adaptation to the gastrointestinal tracts of axenic mice.Appl Environ Microbiol. 2010 May;76(9):2932-9. doi: 10.1128/AEM.02451-09. Epub 2010 Mar 5. Appl Environ Microbiol. 2010. PMID: 20208026 Free PMC article.
-
Bifidobacterium species viability in dairy-based probiotic foods: challenges and innovative approaches for accurate viability determination and monitoring of probiotic functionality.Front Microbiol. 2024 Feb 2;15:1327010. doi: 10.3389/fmicb.2024.1327010. eCollection 2024. Front Microbiol. 2024. PMID: 38371928 Free PMC article. Review.
-
Identification of conserved genomic signatures specific to Bifidobacterium species colonising the human gut.3 Biotech. 2023 Mar;13(3):97. doi: 10.1007/s13205-023-03492-4. Epub 2023 Feb 24. 3 Biotech. 2023. PMID: 36852175 Free PMC article.
References
-
- Ammor, S., K. Yaakoubi, I. Chevallier, and E. Dufour. 2004. Identification by fluorescence spectroscopy of lactic acid bacteria isolated from a small-scale facility producing traditional dry sausages. J. Microbiol. Methods 59:271-281. - PubMed
-
- Araya, M., L. Morelli, G. Reid, M. E. Sanders, and C. Stanton. 2002. Guidelines for the evaluation of probiotics in foods. FAO/WHO report. Food and Agriculture Organization of the United Nations and World Health Organization, Geneva, Switzerland. http://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

