Novel whole-cell antibiotic biosensors for compound discovery
- PMID: 17720843
- PMCID: PMC2075059
- DOI: 10.1128/AEM.00586-07
Novel whole-cell antibiotic biosensors for compound discovery
Abstract
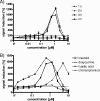
Cells containing reporters which are specifically induced via selected promoters are used in pharmaceutical drug discovery and in environmental biology. They are used in screening for novel drug candidates and in the detection of bioactive compounds in environmental samples. In this study, we generated and validated a set of five Bacillus subtilis promoters fused to the firefly luciferase reporter gene suitable for cell-based screening, enabling the as yet most-comprehensive high-throughput diagnosis of antibiotic interference in the major biosynthetic pathways of bacteria: the biosynthesis of DNA by the yorB promoter, of RNA by the yvgS promoter, of proteins by the yheI promoter, of the cell wall by the ypuA promoter, and of fatty acids by the fabHB promoter. The reporter cells mainly represent novel antibiotic biosensors compatible with high-throughput screening. We validated the strains by developing screens with a set of 14,000 pure natural products, representing a source of highly diverse chemical entities, many of them with antibiotic activity (6% with anti-Bacillus subtilis activity of </=25 mug/ml]). Our screening approach is exemplified by the discovery of classical and novel DNA synthesis and translation inhibitors. For instance, we show that the mechanistically underexplored antibiotic ferrimycin A1 selectively inhibits protein biosynthesis.
Figures

Similar articles
-
[Metabolic products of microorganisms. 62. The inhibition of the incorporation of threonine in sRNA in cell-free systems and of the synthesis of proteins and nucleic acids in the cell by the antibiotic borrelidin].Arch Mikrobiol. 1968;61(2):143-53. Arch Mikrobiol. 1968. PMID: 4974079 German. No abstract available.
-
Application of a Bacillus subtilis Whole-Cell Biosensor (PliaI-lux) for the Identification of Cell Wall Active Antibacterial Compounds.Methods Mol Biol. 2017;1520:121-131. doi: 10.1007/978-1-4939-6634-9_7. Methods Mol Biol. 2017. PMID: 27873249
-
Autoregulation of subtilin biosynthesis in Bacillus subtilis: the role of the spa-box in subtilin-responsive promoters.Peptides. 2004 Sep;25(9):1415-24. doi: 10.1016/j.peptides.2003.11.025. Peptides. 2004. PMID: 15374645
-
The applied side of antimicrobial peptide-inducible promoters from Firmicutes bacteria: expression systems and whole-cell biosensors.Appl Microbiol Biotechnol. 2016 Jun;100(11):4817-29. doi: 10.1007/s00253-016-7519-3. Epub 2016 Apr 22. Appl Microbiol Biotechnol. 2016. PMID: 27102123 Review.
-
Applications of whole-cell bacterial sensors in biotechnology and environmental science.Appl Microbiol Biotechnol. 2007 Jan;73(6):1251-8. doi: 10.1007/s00253-006-0718-6. Epub 2006 Nov 17. Appl Microbiol Biotechnol. 2007. PMID: 17111136 Review.
Cited by
-
A Small Molecule Inhibitor of CTP Synthetase Identified by Differential Activity on a Bacillus subtilis Mutant Deficient in Class A Penicillin-Binding Proteins.Front Microbiol. 2020 Aug 26;11:2001. doi: 10.3389/fmicb.2020.02001. eCollection 2020. Front Microbiol. 2020. PMID: 32973723 Free PMC article.
-
Overview on Strategies and Assays for Antibiotic Discovery.Pharmaceuticals (Basel). 2022 Oct 21;15(10):1302. doi: 10.3390/ph15101302. Pharmaceuticals (Basel). 2022. PMID: 36297414 Free PMC article. Review.
-
Bacillus subtilis extracytoplasmic function (ECF) sigma factors and defense of the cell envelope.Curr Opin Microbiol. 2016 Apr;30:122-132. doi: 10.1016/j.mib.2016.02.002. Epub 2016 Feb 20. Curr Opin Microbiol. 2016. PMID: 26901131 Free PMC article. Review.
-
Quantitative bioassay to identify antimicrobial drugs through drug interaction fingerprint analysis.Sci Rep. 2017 Feb 16;7:42644. doi: 10.1038/srep42644. Sci Rep. 2017. PMID: 28205640 Free PMC article.
-
Curbing gastrointestinal infections by defensin fragment modifications without harming commensal microbiota.Commun Biol. 2021 Jan 8;4(1):47. doi: 10.1038/s42003-020-01582-0. Commun Biol. 2021. PMID: 33420317 Free PMC article.
References
-
- Alksne, L. E., P. Burgio, W. Hu, B. Feld, M. P. Singh, M. Tuckman, P. J. Petersen, P. Labthavikul, M. McGlynn, L. Barbieri, L. McDonald, P. Bradford, R. G. Dushin, D. Rothstein, and S. J. Projan. 2000. Identification and analysis of bacterial protein secretion inhibitors utilizing a SecA-LacZ reporter fusion system. Antimicrob. Agents Chemother. 44:1418-1427. - PMC - PubMed
-
- Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115-119. - PubMed
-
- Au, N., E. Kuester-Schoeck, V. Mandava, L. E. Bothwell, S. P. Canny, K. Chachu, S. A. Colavito, S. N. Fuller, E. S. Groban, L. A. Hensley, T. C. O'Brien, A. Shah, J. T. Tierney, L. L. Tomm, T. M. O'Gara, A. I. Goranov, A. D. Grossman, and C. M. Lovett. 2005. Genetic composition of the Bacillus subtilis SOS system. J. Bacteriol. 187:7655-7666. - PMC - PubMed
-
- Braun, V. 2005. Bacterial iron transport related to virulence. Contrib. Microbiol. 12:210-233. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

