The neutral coalescent process for recent gene duplications and copy-number variants
- PMID: 17720930
- PMCID: PMC2034660
- DOI: 10.1534/genetics.107.074948
The neutral coalescent process for recent gene duplications and copy-number variants
Abstract
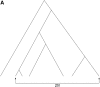
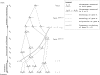
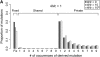
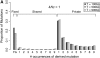
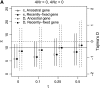
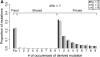
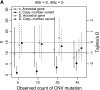
I describe a method for simulating samples from gene families of size two under a neutral coalescent process, for the case where the duplicate gene either has fixed recently in the population or is still segregating. When a duplicate locus has recently fixed by genetic drift, diversity in the new gene is expected to be reduced, and an excess of rare alleles is expected, relative to the predictions of the standard coalescent model. The expected patterns of polymorphism in segregating duplicates ("copy-number variants") depend both on the frequency of the duplicate in the sample and on the rate of crossing over between the two loci. When the crossover rate between the ancestral gene and the copy-number variant is low, the expected pattern of variability in the ancestral gene will be similar to the predictions of models of either balancing or positive selection, if the frequency of the duplicate in the sample is intermediate or high, respectively. Simulations are used to investigate the effect of crossing over between loci, and gene conversion between the duplicate loci, on levels of variability and the site-frequency spectrum.
Figures
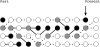










Similar articles
-
Role of selection in fixation of gene duplications.J Theor Biol. 2006 Mar 21;239(2):141-51. doi: 10.1016/j.jtbi.2005.08.033. Epub 2005 Oct 20. J Theor Biol. 2006. PMID: 16242725 Review.
-
Coalescent size versus coalescent time with strong selection.Bull Math Biol. 2007 Oct;69(7):2249-59. doi: 10.1007/s11538-007-9218-9. Epub 2007 Jun 2. Bull Math Biol. 2007. PMID: 17546476
-
Duplicate retention in signalling proteins and constraints from network dynamics.J Evol Biol. 2010 Nov;23(11):2410-21. doi: 10.1111/j.1420-9101.2010.02101.x. Epub 2010 Sep 6. J Evol Biol. 2010. PMID: 20825548
-
Allelic divergence precedes and promotes gene duplication.Evolution. 2006 May;60(5):881-92. Evolution. 2006. PMID: 16817530
-
On selecting markers for association studies: patterns of linkage disequilibrium between two and three diallelic loci.Genet Epidemiol. 2003 Jan;24(1):57-67. doi: 10.1002/gepi.10217. Genet Epidemiol. 2003. PMID: 12508256 Review.
Cited by
-
Molecular evolution of the three short PGRPs of the malaria vectors Anopheles gambiae and Anopheles arabiensis in East Africa.BMC Evol Biol. 2010 Jan 12;10:9. doi: 10.1186/1471-2148-10-9. BMC Evol Biol. 2010. PMID: 20067637 Free PMC article.
-
Both positive and negative selection pressures contribute to the polymorphism pattern of the duplicated human CYP21A2 gene.PLoS One. 2013 Nov 29;8(11):e81977. doi: 10.1371/journal.pone.0081977. eCollection 2013. PLoS One. 2013. PMID: 24312389 Free PMC article.
-
Interplay of interlocus gene conversion and crossover in segmental duplications under a neutral scenario.G3 (Bethesda). 2014 Jun 6;4(8):1479-89. doi: 10.1534/g3.114.012435. G3 (Bethesda). 2014. PMID: 24906640 Free PMC article.
-
Unified modeling of gene duplication, loss, and coalescence using a locus tree.Genome Res. 2012 Apr;22(4):755-65. doi: 10.1101/gr.123901.111. Epub 2012 Jan 23. Genome Res. 2012. PMID: 22271778 Free PMC article.
-
Chimeric genes as a source of rapid evolution in Drosophila melanogaster.Mol Biol Evol. 2012 Feb;29(2):517-29. doi: 10.1093/molbev/msr184. Epub 2011 Jul 18. Mol Biol Evol. 2012. PMID: 21771717 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources

