Mutational interference and the progression of Muller's ratchet when mutations have a broad range of deleterious effects
- PMID: 17720933
- PMCID: PMC2034659
- DOI: 10.1534/genetics.107.073791
Mutational interference and the progression of Muller's ratchet when mutations have a broad range of deleterious effects
Abstract
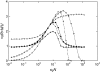
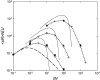
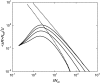
Deleterious mutations can accumulate in asexual haploid genomes through the process known as Muller's ratchet. This process has been described in the literature mostly for the case where all mutations are assumed to have the same effect on fitness. In the more realistic situation, deleterious mutations will affect fitness with a wide range of effects, from almost neutral to lethal. To elucidate the behavior of the ratchet in this more realistic case, simulations were carried out in a number of models, one where all mutations have the same effect on selection [one-dimensional (1D) model], one where the deleterious mutations can be divided into two groups with different selective effects [two-dimensional (2D) model], and finally one where the deleterious effects are distributed. The behavior of these models suggests that deleterious mutations can be classified into three different categories, such that the behavior of each can be described in a straightforward way. This makes it possible to predict the ratchet rate for an arbitrary distribution of fitness effects using the results for the well-studied 1D model with a single selection coefficient. The description was tested and shown to work well in simulations where selection coefficients are derived from an exponential distribution.
Figures






Similar articles
-
The advance of Muller's ratchet in a haploid asexual population: approximate solutions based on diffusion theory.Genet Res. 1993 Jun;61(3):225-31. doi: 10.1017/s0016672300031384. Genet Res. 1993. PMID: 8365659
-
The traveling-wave approach to asexual evolution: Muller's ratchet and speed of adaptation.Theor Popul Biol. 2008 Feb;73(1):24-46. doi: 10.1016/j.tpb.2007.10.004. Epub 2007 Oct 22. Theor Popul Biol. 2008. PMID: 18023832 Free PMC article.
-
The speed of Muller's ratchet with background selection, and the degeneration of Y chromosomes.Genet Res. 2001 Oct;78(2):149-61. doi: 10.1017/s0016672301005213. Genet Res. 2001. PMID: 11732092
-
The Muller's Ratchet and Aging.Trends Genet. 2020 Jun;36(6):395-402. doi: 10.1016/j.tig.2020.02.004. Epub 2020 Mar 18. Trends Genet. 2020. PMID: 32396833 Review.
-
[Evolution of sex: role of deleterious mutation and mobile elements].Zh Obshch Biol. 2003 Nov-Dec;64(6):463-78. Zh Obshch Biol. 2003. PMID: 14723170 Review. Russian.
Cited by
-
Muller's ratchet and the degeneration of the Drosophila miranda neo-Y chromosome.Genetics. 2010 May;185(1):339-48. doi: 10.1534/genetics.109.112789. Epub 2010 Mar 9. Genetics. 2010. PMID: 20215466 Free PMC article.
-
The effects of recombination rate on the distribution and abundance of transposable elements.Genetics. 2008 Apr;178(4):2169-77. doi: 10.1534/genetics.107.082743. Genetics. 2008. PMID: 18430942 Free PMC article.
-
On the potential for extinction by Muller's ratchet in Caenorhabditis elegans.BMC Evol Biol. 2008 Apr 30;8:125. doi: 10.1186/1471-2148-8-125. BMC Evol Biol. 2008. PMID: 18447910 Free PMC article.
-
Kick-starting the ratchet: the fate of mutators in an asexual population.Genetics. 2011 Apr;187(4):1129-37. doi: 10.1534/genetics.110.124818. Epub 2011 Feb 1. Genetics. 2011. PMID: 21288878 Free PMC article.
-
Population dynamics of PIWI-interacting RNAs (piRNAs) and their targets in Drosophila.Genome Res. 2010 Feb;20(2):212-27. doi: 10.1101/gr.095406.109. Epub 2009 Nov 30. Genome Res. 2010. PMID: 19948818 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

